Atomic Positioning with DNA Hinges
Table of Contents
DNA Hinge Key Points
- Funke and Dietz created a DNA hinge which is adjustable by varying the length of one of the DNA molecules, allowing positioning of molecules attached to the hinge by as little as 0.04 nanometers.
- DNA origami was one of the first successful methods of “bottom–up” nanotechnology, allowing structures such as cubes, smiley faces, and boxes with functional lids to be designed.
- DNA has the advantage over other biological polymers like proteins and RNA that the structure and position of every atom can be predicted before assembly.
- Funke and Dietz used DNA origami to precisely control the distance between two molecules, measuring the difference in FRET intensity of molecules separated from 1.5 to 9 nanometers in 123 steps of as little as 0.4 angstroms each.
- Applications of this research include templating chemical reactions at precise locations, probing interatomic forces at precisely defined distances, and mapping out the structure of proteins.
I was perusing the Nature Nanotechnology website today when I came across an interesting article by Funke and Dietz, called “Placing molecules with Bohr radius resolution using DNA origami.” The authors created a hinge out of DNA which they selectively open or close by varying the length of one of the DNA molecules, which allows them to adjust the positions of molecules attached to the hinge by as little as 0.04 nanometers, much less than the typical spacing between atoms in a solid.
Machines Made of Atoms
One of the initial visions of nanotechnology, in Richard Feynman’s essay “There’s Plenty of Room at the Bottom,” was that tiny machines could be built atom by atom, arranged according to our design. This, however, is not the way the field of nanotechnology progressed: most of its successes revolved around making smaller and smaller electronic components using “top-down” procedures such as electron-beam lithography. This was very useful, but not akin to the idea of “bottom-up” nanotechnology that was dreamed about. One of the first successful methods of bottom-up nanotechnology was the advent of DNA origami in the mid-2000s. In Rothemund’s 2006 paper, he combined a long strand of viral DNA with many short “staple” strands that he designed himself, which bound to the longer strand in such a way as to make it self-assemble into, among other things, 100-nanometer smiley faces. The ability to design these structures has progressed since then, with 3D structures and boxes that can open and close. Originally this seemed like a solution without a problem, but it has found applications in drug delivery (e.g. a box that only opens and releases its content if it is unlocked by the right gene) and scaffolding for arranging other things onto DNA lattices for top-down lithography. Here is a video demonstrating how these things self-assemble.
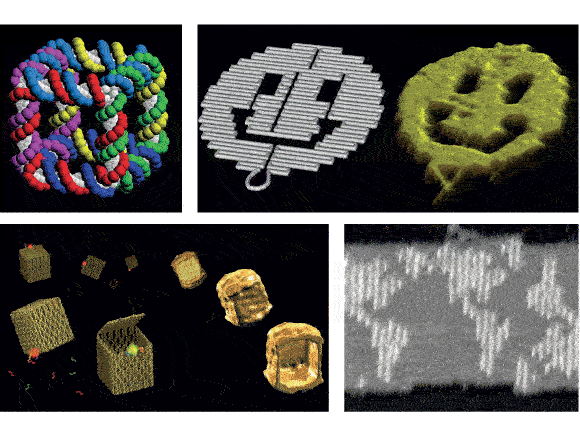
Some examples of DNA origami: are a cube, a smiley face, a box with a functional lid, and a map. Source.
DNA Machines
Using DNA has the advantage over other biological polymers like proteins and RNA that we can completely dictate and understand the structure through the choice of the strand sequence. This means that the position of every base can be predicted before the structure is assembled, and within that, the position of each atom can be dictated. Atoms can also be moved around one at a time with scanning probe microscopes, but this is extremely slow, limited to two dimensions, and not mass-producible in the way that chemical self-assembly is. Funke and Dietz were interested in using DNA origami to specifically control the distance between two molecules. The standard approach to that type of thing involved creating a “breadboard” in a lattice of DNA origami and positioning things within the lattice. They wanted to do better.
They created what essentially amounts to an adjustable DNA hinge: two comparatively rigid backbones made of DNA origami structures, with a triangle of DNA double-helices controlling the interior angle of the two structures. By dictating the number of base pairs in the supporting leg of the triangle, they could control the opening angle. By adjusting it one base-pair at a time, from 10 to 50 base-pairs, roughly 3 to 17 nanometers (a good rule of thumb is that double-stranded DNA has three base-pairs per nanometer), they adjusted in the opening angle of the hinge in 41 discrete steps. Looking at the hinges with a transmission electron microscope, it is clear that the angle can be precisely controlled.
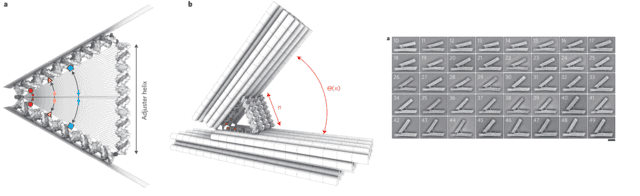
Images from the paper showing the design of the hinge (the cylinders are made of DNA as well). Electron microscopy images show it opening at different angles based on the number of base pairs in the opening strand. The scale bar is 20 nanometers.
To see how well they could position molecules using this hinge, they attached two fluorescent molecules to the interior arms of the hinge triangle, in equivalent positions (the colored dots in the above picture). These molecules undergo Förster resonant energy transfer (FRET), a phenomenon where optically excited molecules transfer their excess energy to unexcited molecules through dipole forces. Through this mechanism, the originally unexcited molecule emits light that is a lower frequency than the excitation of the first molecule, so by detecting this lower frequency emission it can be ascertained that one molecule transferred its energy to another. These dipole interactions are extremely short-range and decay with the sixth power of distance, it is used in biophysics and related fields to see whether molecules come into physical contact with one another (to the extent that that is meaningful on the atomic scale), and exactly how close they get.

Diagram of how FRET works. The important thing is that it only works if the molecules are really close. Source.
So, Funke and Dietz measured the FRET intensity coming out of these marker molecules from hinges with different opening angles. They had two independent estimates of the distance: the FRET intensity, and their knowledge of the hinge’s structure. By comparing them, they found out, unsurprisingly, that the FRET intensity is correlated with the structural position of the dye molecules. They could accurately measure the difference in FRET intensity of molecules separated from 1.5 to 9 nanometers, in 123 (41 x 3) steps of as little as 0.4 angstroms each. That is an extremely precise measurement, one of the most precise I know of in condensed matter physics. As they state in the paper, this is comparable to the Bohr radius of an atom, the average distance from the nucleus to the first electron. Atoms in a solid are typically separated by a few angstroms, and they can position molecules on these DNA hinges with much greater precision than that: this is atomic positioning with sub-atomic resolution.
In the “How this will change the world if we get more money” section that concludes every paper, the authors talk about how this will lead to new applications of DNA origami to templating chemical reactions at precise locations, to probe interatomic forces at precisely defined distances, and to map out the structure of proteins by binding them to specific sites on these hinges, essentially creating the worlds finest ruler. All of these are pretty cool ideas.
Conclusion
I chose to do my graduate research on using top-down nanotechnology to study DNA in part because I thought DNA origami, which was only a few years old at the time, was cool. One aspect of my master’s project was about controlling what structure DNA will fold into at equilibrium, although my Ph.D. was more quantitative. Checking up on the field every few years, it’s impressive to see the progress that this field is making, and every time I read a DNA origami paper I learn something interesting. I look forward to seeing where it goes in the next few years.
Ph.D. McGill University, 2015
My research is at the interface of biological physics and soft condensed matter. I am interested in using tools provided from biology to answer questions about the physics of soft materials. In the past I have investigated how DNA partitions itself into small spaces and how knots in DNA molecules move and untie. Moving forward, I will be investigating the physics of non-covalent chemical bonds using “DNA chainmail” and exploring non-equilibrium thermodynamics and fluid mechanics using protein gels.



I wasn’t thinking of positioning of atoms as in to piece together parts, like on a workbench. I was thinking of the DNA backbone as the “bench” to assemble pieces within itself, similar to protein mechanics. I am quite fuzzy on what actually is state of the “art”, as Greg called it… I think there might be a lot more going on behind the scenes or there should be! I can only imagine how much potential this field has and I can’t make out many challenges it couldn’t help overcome.
Three statisticians are playing darts in a bar. The first steps up to the line, throws his dart and misses wide left. The second statistician takes his turn, and misses wide right, at which point the third statistician yells “bullseye!”
After having time to read the full paper, this is my impression of the claim that the authors can position molecules with “Bohr radius” accuracy. Yes, they can construct structures where they molecules with an accuracy of 0.04 nm, but the precision with which they can place the molecules is an order of magnitude larger (~0.5 nm from the authors’ estimates). The result is still impressive and useful, though claims of Bohr radius resolution may be a bit overblown.
“I haven’t seen anything more recent on the topic.”
Me either. Anything in this realm attracts my interest! Having such fine angle control the first thing I’d want to make is atomic pinwheels to see if we can figure out how to make it do something…
“The article focuses mostly on supramolecular chemistry, though one could imagine trying to design DNA nanostructures capable of such functions as well.”
They’re already beyond mere imagination, though I haven’t seen anything more recent on the topic.
[URL]http://www.nature.com/nature/journal/v459/n7243/full/nature07971.html[/URL]
“I could see DIY kits out by Christmas :biggrin:”
I was thinking the same thing! I just couldn’t think of anything other than nanorobotic drain opener, and I doubt that’s on anyone but the plumber’s x-mas list.
Oooh, nanorobotic paint scrapers, too! I could use a few bucketfuls of those. :eek: I suppose they should collect lead as well…
Why can’t I think of anything fun…
“It makes me want to build nanobots!”
I could see DIY kits out by Christmas :biggrin:
“This is going to be a new category in physics soon? Nano-mechanics? It makes me want to build nanobots!”
You’re not the only one interested in nanobots: [URL]https://www.physicsforums.com/threads/making-a-voltron-out-of-dna.807896/[/URL] (of course, the DNA origami nanobot in that article only looks like a robot, but doesn’t actually do anything). Here’s a news piece from [i]Nature[/i] discussion chemists efforts to build nanoscale parts to generate molecular scale motion: [URL]http://www.nature.com/news/the-tiniest-lego-a-tale-of-nanoscale-motors-rotors-switches-and-pumps-1.18262[/URL] The article focuses mostly on supramolecular chemistry, though one could imagine trying to design DNA nanostructures capable of such functions as well.
The possible places this could lead to in 20 years blow my mind. Hope I’m around to see it developed.
Dear staffi wowed alot about this topicthanks alot
This is going to be a new category in physics soon? Nano-mechanics? It makes me want to build nanobots!
Wow! The birth of a new art?