Why CRISPR Technologies Won’t Lead to Designer Babies
 The prospect of designer babies has been a staple of the dystopian futures imagined by science-fiction writers, evoking fears of eugenics (e.g. Brave New World) or social inequality (e.g. Gattaca). However, new advances in gene editing technologies, in particular, the discovery of the CRISPR-Cas9 system, are bringing this piece of science fiction closer to reality. Last week, researchers published the first study demonstrating gene editing in human embryos. More studies are likely to come as universities and companies around the world pursue the same goal for a variety of scientific and clinical applications. Despite calls from some in the scientific community to place a moratorium on gene editing in embryos, such a moratorium would be difficult to enforce worldwide. What are the capabilities of this new technology and what might it hold for our future? In this Insight, I will address these questions by discussing the science behind gene editing, its limitations, and the future of gene editing in the clinic.
The prospect of designer babies has been a staple of the dystopian futures imagined by science-fiction writers, evoking fears of eugenics (e.g. Brave New World) or social inequality (e.g. Gattaca). However, new advances in gene editing technologies, in particular, the discovery of the CRISPR-Cas9 system, are bringing this piece of science fiction closer to reality. Last week, researchers published the first study demonstrating gene editing in human embryos. More studies are likely to come as universities and companies around the world pursue the same goal for a variety of scientific and clinical applications. Despite calls from some in the scientific community to place a moratorium on gene editing in embryos, such a moratorium would be difficult to enforce worldwide. What are the capabilities of this new technology and what might it hold for our future? In this Insight, I will address these questions by discussing the science behind gene editing, its limitations, and the future of gene editing in the clinic.
Table of Contents
How editing a gene works
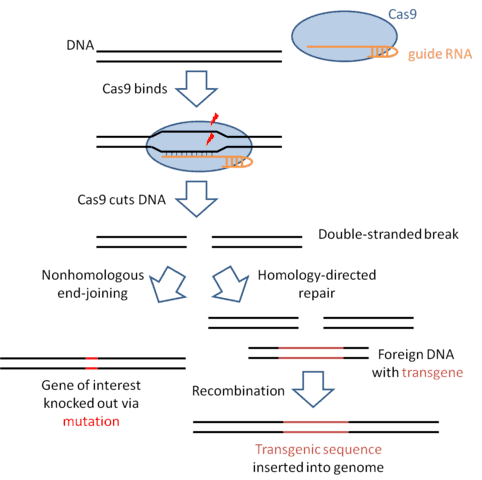
The principle behind targeted gene editing is fairly straightforward. If you create a double-stranded break in DNA, this break can be repaired in one of two ways (see the diagram below). One way, called nonhomologous end-joining, involves stitching the two broken pieces of DNA together, often creating a mutation at the site where the DNAs are rejoined. Therefore, if you cut a protein-coding DNA sequence, more often than not, you’ll introduce a mutation that will knock out the function of that gene. The second way in which the cell can repair a double-stranded break is a process called homology-directed repair. Here, the repair machinery makes use of the fact that we carry two copies of our genome, one which we inherit from our father and one which we inherit from our mother. The machinery searches out the matching DNA sequence on the undamaged copy of the chromosome (the homologous chromosome), and then copies that sequence onto the damaged chromosome. For genome engineering purposes, we can trick the cell by adding in a foreign DNA molecule that looks similar to the region that we cut. Instead of grabbing the homologous chromosome, the repair machinery will grab the foreign DNA and copy that (along with whichever transgenic sequences we want to add to the genome) onto the damaged chromosome. So all you need to do to edit a gene is find a way to cut the genome at a specific sequence, and the cell’s DNA repair machinery will take care of the rest for you.
If the principle behind gene editing is so simple, why wasn’t it done earlier? The bottleneck in the process has been finding a general method to cut the DNA at a specified sequence. Early efforts in this area involved engineering proteins to recognize specific DNA sequences, but this process turned out to be fairly difficult, often requiring a few rounds of testing and redesign to get a protein that targets the right sequence. Thus, anytime you wanted to target a new DNA sequence you’d have to begin a laborious effort to design a new protein. The discovery of the CRISPR-Cas9 system changed all of that. This system, which comes from the antiviral defense mechanisms used by bacteria, consists of two parts: the Cas9 protein which houses the enzyme active site that cuts the DNA, and a guide RNA (in bacteria, this RNA comes from a region of the genome containing clustered regularly interspaced short palindromic repeats, hence the acronym CRISPR). The guide RNA contains a 20 bp guide sequence that tells Cas9 which sequence to cut. Because the guide sequence works through simple Watson-Crick base pairing, designing a new cutting enzyme is simple and requires changing only the 20 bp guide sequence (as well as making sure that the target sequence contains a small sequence motif recognized by the Cas9 protein). The ease with which Cas9 can be targeted to any arbitrary sequence makes CRISPR-mediated gene editing available to anyone with reasonable training in molecular biology.
The limitations of gene editing
So now that we can edit the human genome at will, super-intelligent babies must be just around the corner, right? Unfortunately, the genetics of many traits that we care about are quite complex and difficult to manipulate. Take the genetics of human height, for example. We know from previous studies that genetics explains ~ 80% of the variation in height among individuals, with environmental effects (differing diets, etc.) accounting for the other 20%. Furthermore, researchers have identified many of the genes responsible for determining an individual’s height. The problem, however, is that each of these genes individually has a tiny effect on height. For example, it takes the combined effect of ~700 genetic variants to account for ~20% of the variation in human height. So unless you’re editing hundreds to thousands of genes at a time, you’ll only minutely affect the chance of your baby being tall. This is likely true for other complex traits, like intelligence.
This discussion, of course, presupposes that we understand which changes we’d like to make to the genome. The studies above that identified the thousands of genes that influence human height were found in so-called genome-wide association studies (GWAS). The keyword here is association: these studies identified genetic variants associated with human height, but the studies cannot determine whether the variants are causing the differences in human height. Many of these variants probably do not influence human height themselves, but instead, co-segregate with a nearby variant that influences human height. Unfortunately, knowing how to edit genomes does not mean we know what to edit.
Of course, there are examples of traits whose genetics is much simpler. Many genetic disorders, such as sickle-cell anemia, cystic fibrosis, and Huntington’s disease, are caused by well-characterized changes to a single gene that we know to be causative of these disorders. Gene editing offers the prospect of repairing the mutations that cause these single-gene disorders. While research is moving forward to use CRISPR to treat individuals with genetic disorders, gene editing is not required if the goal is to engineer embryos free from genetic disorders. In most cases, doctors can instead sequence the DNA from embryos to determine which embryos would have a genetic disorder and which would produce healthy children. The doctors can then use only the healthy embryos for in vitro fertilization. Sequencing and screening embryos, called preimplantation genetic diagnosis, is a much easier, safer, and cheaper method than using gene editing to achieve the same goal.
Are there any useful applications of gene editing in embryos? It turns out that there are a few rare cases of beneficial traits conferred largely by a single gene. For example, researchers have identified individuals who lack functional copies of the gene PCSK9, a gene involved in cholesterol metabolism, who are perfectly normal except that they have unusually low amounts of bad cholesterol, suggesting that they are at lower risk for cardiovascular disease. Individuals who carry mutations in the CCR5 protein, one of the co-receptors for HIV, are immune to HIV infection. A particular mutation in the APP protein, the protein that forms plaques in the brains of those with Alzheimer’s disease, lowers one’s risk of Alzheimer’s disease. Gene editing offers the ability to introduce these rare mutations into embryos to lower their lifetime risk of cardiovascular disease or Alzheimer’s or confer resistance to HIV infection. Of course, the standard of safety for these types of modifications is much higher than in therapeutic applications: For therapies that intend to fix a genetic disorder, side effects from the procedure are tolerable as long as they are not worse than the disease they’re curing. For therapies intended to treat asymptomatic individuals, even mild side effects can skew the cost-benefit ratio.
So far, the list of such beneficial traits is small, but as full genome sequencing becomes more commonplace, perhaps we will identify more beneficial single-gene traits. Ultimately, we can find such traits that will limit the possibilities of gene editing to create genetically enhanced humans.
Are we headed for a brave new world with CRISPR?
So, what does this new technology mean for the future of medicine? CRISPR will continue to be an important tool for biomedical research, allowing researchers to more easily generate new cell lines or animal models to study human disease. For use in the clinic, either for gene therapy in individuals with genetic disorders or for the modification of embryos, CRISPR and other gene-editing technologies still face many challenges regarding their safety and efficacy. For example, CRISPR cuts fairly efficiently at its target sequence, but it can also cut at “off-target” sites, introducing unwanted mutations elsewhere in the genome. Figuring out how to reduce off-target effects is a must before CRISPR enters the clinic. Even if these issues are resolved, however, we lack the necessary information to know a priori the effects of most genetic changes we introduce, so research must proceed with caution. Without more fundamental breakthroughs in our understanding of biology, we have only a limited capacity to genetically enhance embryos, so CRISPR will not lead to a Gattaca-like world with a genetically engineered upper class, ruling over a non-engineered lower class anytime soon. Hopefully, CRISPR will instead find its first clinical uses in therapeutic applications like helping those with genetic diseases or engineering immune cells to help fight HIV or cancer.
Further CRISPR Reading:
Hsu, Lander & Zhang 2014. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157: 1262
Regaldo 2015. Engineering the Perfect Baby. MIT Technology Review Magazine.
Postdoctoral researcher in biophysics.
My areas of expertise include single molecule spectroscopy, structural biology, biochemistry, and evolutionary biology.





UK scientists are allowed to test CRISPR-Cas9 with human embryos.
[URL]http://www.bbc.co.uk/news/health-35459054[/URL]
[URL]http://bigstory.ap.org/article/fdda5bf9f0314b748c7438c9659da83a/britain-approves-controversial-gene-editing-technique[/URL]
More news on from those trying to engineer Cas9 proteins with increased fidelity:
Kleinstiver et al. 2016 High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. [i]Nature[/i]. Published online 06 Jan 2016. [URL=’http://dx.doi.org/10.1038/nature16526′]doi:10.1038/nature16526[/URL]
[quote]CRISPR–Cas9 nucleases are widely used for genome editing but can induce unwanted off-target mutations. Existing strategies for reducing genome-wide off-target effects of the widely used Streptococcus pyogenes Cas9 (SpCas9) are imperfect, possessing only partial or unproven efficacies and other limitations that constrain their use. Here we describe SpCas9-HF1, a high-fidelity variant harbouring alterations designed to reduce non-specific DNA contacts. SpCas9-HF1 retains on-target activities comparable to wild-type SpCas9 with >85% of single-guide RNAs (sgRNAs) tested in human cells. Notably, with sgRNAs targeted to standard non-repetitive sequences, SpCas9-HF1 rendered all or nearly all off-target events undetectable by genome-wide break capture and targeted sequencing methods. Even for atypical, repetitive target sites, the vast majority of off-target mutations induced by wild-type SpCas9 were not detected with SpCas9-HF1. With its exceptional precision, SpCas9-HF1 provides an alternative to wild-type SpCas9 for research and therapeutic applications. More broadly, our results suggest a general strategy for optimizing genome-wide specificities of other CRISPR-RNA-guided nucleases.[/quote]
News articles:
Enzyme tweak boosts precision of CRISPR genome edits [URL]http://www.nature.com/news/enzyme-tweak-boosts-precision-of-crispr-genome-edits-1.19114[/URL]
Improved Version Of CRISPR Gene Editing Tool Eliminates Errors [URL]http://www.popsci.com/new-form-crispr-is-more-precise[/URL]
The work seems complementary to the work published by the Zhang lab (mentioned in [URL=’https://www.physicsforums.com/threads/dont-fear-the-crispr-comments.811056/page-2#post-5305836′]post #30[/URL]), so the different modifications can probably be combined to engineer an even more precise enzyme.
“What Happens If Someone Uses This DIY Gene Hacking Kit to Make Mutant Bacteria?
[URL=’http://motherboard.vice.com/read/what-happens-if-someone-uses-this-diy-gene-hacking-kit-to-make-mutant-bacteria’]http://motherboard.vice.com/read/wh…-diy-gene-hacking-kit-to-make-mutant-bacteria[/URL]”
You don’t really need CRISPR to make precision edits to bacteria, and the technology to do this has been around since the late 80s ([URL]https://en.wikipedia.org/wiki/Recombineering[/URL]). The Indiegogo page for the kit in question describes the types of experiments that might be performed in freshman biology courses. These are not dangerous experiments. Yes, in theory, someone could use CRISPR to engineer something potentially illegal (for example, yeast that allow you to [url=http://www.wired.com/2015/08/dont-try-home-scientists-brew-opiates-yeast/]brew illegal drugs[/url]), but doing so would take much more expertise than the general public would have.
Personally, I would be more worried about people being able to stockpile assault weapons and materials for pipe bombs at home.
Not a great source I know, but good to start the conversation
What Happens If Someone Uses This DIY Gene Hacking Kit to Make Mutant Bacteria?
[URL]http://motherboard.vice.com/read/what-happens-if-someone-uses-this-diy-gene-hacking-kit-to-make-mutant-bacteria[/URL]
Like with any new technology, I think I feel a reasonable amount of apprehensiveness, most of which likely comes from the fact that I have absolutely no background in cell biology or genetics and can’t speak to the limitations of this technology one way or another, but I also know that many other technological revolutions have come and gone and the world is a better place for it despite some people at the time being absolutely terrified of them.
I’m hopeful, to say the least.
On the topic of Cas9 fidelity, there were two recent news articles posted today:
[URL]http://www.nature.com/news/biologists-create-more-precise-molecular-scissors-for-genome-editing-1.18932[/URL]
[URL]http://www.theatlantic.com/science/archive/2015/12/who-edits-the-gene-editors/418209/[/URL]
Which are about a [URL=’http://www.sciencemag.org/content/early/2015/11/30/science.aad5227′]Science paper[/URL] from Feng Zhang’s lab published today on the topic.
“[USER=13297]@Yggdrasil[/USER] I think you’re wrong there. There are various methods developed in very recent times which have been shown to increase the fidelity of cleavage.[/quote]
Yes, I was probably unclear here. I meant that the fidelity of a single CRISPR-Cas9 in the standard gene editing protocols is probably insufficient for many clinical applications. Modified versions (such as the engineered Cas9 version described above or the dimeric versions) may end up being useful. Of course, the amount of off target activity that is tolerable will depend a lot on the particular application (e.g. off target activity will be more acceptable when treating serious diseases whereas it will be much less acceptable when editing genes in healthy individuals).
[quote]a Cas9-FokI nuclease combination was shown to reduce off-target effects by requiring dimerization and two gRNA sequences to achieve a DSB. My understanding was that the gRNA still served as the means of targeting the DNA, and the attached FokI domains were used as a nuclease, to replace the nuclease function of dCas9 (de-activated Cas9). FokI simply requires dimerization to act as a nuclease (see “Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing” [URL]http://www.nature.com/nbt/journal/v32/n6/full/nbt.2908.html[/URL]). Within that paper it’s referred to as wild-type FokI nuclease that was attached to dCas9, used on multiple sites.[/quote]
Yes, I linked to this study in my [URL=’https://www.physicsforums.com/threads/dont-fear-the-crispr-comments.811056/page-2#post-5302914′]post #27[/URL]. One problem with this approach is that DNA binding by dCas9 is much more promiscuous than DNA cleavage by catalytically competent Cas9 (see for example [URL=’http://www.nature.com/nbt/journal/v32/n7/full/nbt.2889.html’]this paper from Phil Sharp’s lab[/URL] showing widespread off-target binding of dCas9 compared to Cas9 cutting with the same gRNA and [URL=’http://www.nature.com/nature/journal/v527/n7576/full/nature15544.html’]this paper from Jennifer Doudna’s lab[/URL] providing a mechanistic basis for the additional proofreading that occurs between DNA binding and DNA cleavage). A more promising approach is the other paper I cited from Feng Zhang’s lab that uses a [URL=’http://www.cell.com/abstract/S0092-8674%2813%2901015-5′]double nicking[/URL] approach that likely preserves the fidelity of Cas9 compared to strategies using dCas9.
[quote]My understanding is that the Zinc-finger part of the ZFN is the DNA-binding domain, FokI is simply a nuclease attached to the Zinc-finger protein ([URL]https://en.wikipedia.org/wiki/Zinc_finger_nuclease[/URL]), same with TALENs, TALE is the DNA-binding domain and FokI is simply an attached nuclease. The main problem I thought was the efficacy of cutting and maybe that’s where protein engineering might be needed.[/quote]
No, the main drawback of ZFNs and TALENs is the difficulty in engineering them to target new sequences. See Table 1 in [URL]http://www.nature.com/nm/journal/v21/n2/full/nm.3793.html[/URL] for a comparision of various genome editing technologies.
[quote]Given the availability of the ZF nickase technique (where a pare of FokI dimers is converted to a nickase rather than a nuclease), if you can find further efficiency gains (as I recall efficacy is quite poor even in a Zinc finger-nickase), you could use with Cas9 to the point it would require dimerization to make a single nick – thus 4 gRNA sequences would be required to make a single DSB. You’ve also got the achievement of enhanced specificities through use of tru-gRNA (truncated gRNAs – reduces undesired mutagenesis some 5,000 fold [URL]http://www.nature.com/nbt/journal/v32/n3/full/nbt.2808.html[/URL]), so my conclusion is that if you get these two working in concert with improvements in predictive software, you’re gonna end up with Cas9 of quite a high targeting specificity.”
At some point, with Cas9 you will begin having problems finding suitable target sites due to the invariant PAM recognition site of Cas9. At some targets you will not have a suitable number of PAM sites positioned correctly to use dimeric or tetrameric Cas9 strategies, so ZFNs or TALENs may be required for these targets. There have been some successes in [url=www.nature.com/nature/journal/v523/n7561/abs/nature14592.html]reengineering Cas9 to recognize other PAM sequences[/url], however, so these advances could remove some of these limitations.
[USER=13297]@Yggdrasil[/USER] I think you’re wrong there. There are various methods developed in very recent times which have been shown to increase the fidelity of cleavage. I covered a lot of them, plus potential developments for the future when I was carrying out a risk assessment of whether Cas9 is safe to use in human beings or for widescale release to the environment. When I covered it a Cas9-FokI nuclease combination was shown to reduce off-target effects by requiring dimerization and two gRNA sequences to achieve a DSB. My understanding was that the gRNA still served as the means of targeting the DNA, and the attached FokI domains were used as a nuclease, to replace the nuclease function of dCas9 (de-activated Cas9). FokI simply requires dimerization to act as a nuclease (see “Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing” [URL]http://www.nature.com/nbt/journal/v32/n6/full/nbt.2908.html[/URL]). Within that paper it’s referred to as wild-type FokI nuclease that was attached to dCas9, used on multiple sites.
My understanding is that the Zinc-finger part of the ZFN is the DNA-binding domain, FokI is simply a nuclease attached to the Zinc-finger protein ([URL]https://en.wikipedia.org/wiki/Zinc_finger_nuclease[/URL]), same with TALENs, TALE is the DNA-binding domain and FokI is simply an attached nuclease. The main problem I thought was the efficacy of cutting and maybe that’s where protein engineering might be needed.
Given the availability of the ZF nickase technique (where a pare of FokI dimers is converted to a nickase rather than a nuclease), if you can find further efficiency gains (as I recall efficacy is quite poor even in a Zinc finger-nickase), you could use with Cas9 to the point it would require dimerization to make a single nick – thus 4 gRNA sequences would be required to make a single DSB. You’ve also got the achievement of enhanced specificities through use of tru-gRNA (truncated gRNAs – reduces undesired mutagenesis some 5,000 fold [URL]http://www.nature.com/nbt/journal/v32/n3/full/nbt.2808.html[/URL]), so my conclusion is that if you get these two working in concert with improvements in predictive software, you’re gonna end up with Cas9 of quite a high targeting specificity.
“A major focus of research on gene editing is increasing the fidelity of cleavage by the CRISPR-Cas9 system.”
Sounds like we need an update Insight :wink::smile:
“[USER=124113]@Ygggdrasil[/USER]: Can you comment on [URL=’http://www.sciencedaily.com/releases/2015/11/151118155446.htm’]this news[/URL]? It looks very interesting.”
A major focus of research on gene editing is increasing the fidelity of cleavage by the CRISPR-Cas9 system. The Cas9 enzyme can tolerate a number of mismatches in its target site, so it is prone to cutting at sites that resemble the target site. These off-target cleavages can be problematic as they can lead to unwanted edits to the genome that could be problematic, especially in clinical applications (in the worst case scenario, an off target mutation could lead to a serious disease like cancer). The news article (which references [url=http://www.nature.com/nmeth/journal/vaop/ncurrent/full/nmeth.3624.html]this paper[/url] from the journal [i]Nature Methods[/i]) discusses a new method that combines elements from two different gene editing approaches to increase the potential for off target effects of gene editing.
Prior to the discovery of the CRISPR-Cas9 system, researchers had been working with two different technologies, zinc-finger (ZFs) and tal effector (TALEs), as ways to perform gene editing. These are proteins which can be engineered to recognize and cut specific sequences in the human genome to perform gene editing. The advantage they have over CRISPR-Cas9 is that they are generally better at distinguishing the correct sequence from incorrect sequences and are less likely to introduce unwanted edits at other sites in the genome. However, it is relatively difficult to engineer new ZFs or TALEs to recognize new sequences. In contrast, it is trivial to program the CRISPR-Cas9 to target new sequences, but as mentioned above, suffers from problems with off target cleavage.
To increase the fidelity of the CRISPR-Cas9 system, the authors add either a ZF or a TALE as a DNA-binding domain (DBD) to a weakened version of the Cas9 enzyme. The weakened Cas9 is not capable of binding to its target site on its own, but instead relies on the DBD for recruitment. Once tethered near its target site by the DBD, the Cas9 can then read its target sequence and cleave the DNA for gene editing purposes. Because cleavage depends on recognition of both the binding site for the DBD and the guide RNA of the CRISPR system, the engineered protein shows increased fidelity. This strategy also helps expand the types of targets that CRISPR can recognize.
The main drawback here is that the technique relies on ZFs and TALEs, and as mentioned previously, it is difficult to reprogram ZFs or TALEs to recognize new sequences. The authors of this study targeted a site in the genome that they have studied extensively for which ZFs and TALEs had previously been designed (and these ZFs and TALEs have been well validated in the literature). Designing a new construct to target a new site would involve much more work to redesign and test new ZFs or TALEs. In contrast, other methods exist for increasing the fidelity of the CRISPR-Cas9 system, which does not sacrifice the ease of reprogramming the nucleases to recognize new sequences (for example, see [URL]http://www.nature.com/nbt/journal/v32/n6/full/nbt.2908.html[/URL] or [URL]http://www.cell.com/abstract/S0092-8674%2813%2901015-5[/URL]). The authors of the [i]Nature Methods[/i] study claim, ~100-fold reduction of off target cleavage, whereas the double-nicking strategy in the [i]Cell[/i] paper claims a 50-1000 fold reduction in off-target cleavage, so it’s not clear that the method is better than other existing methods. There may be some contexts, however, where the DBD-fusion strategy will work where some of the other strategies might not work.
While the ease of programming CRISPR-Cas9 recognition will be useful in basic research for screening efforts, it’s likely that the fidelity of the CRISPR-Cas9 system will not be sufficient for clinical applications. Clinical applications will probably still require testing many different approaches (CRISPR-based, ZF-based, TALE-based, and various combinations) in order to find the approach that leads to the least off target effects. Whether one approach is superior in all cases or whether the best approach depends on the particular target and application likely remains to be seen.
[USER=124113]@Ygggdrasil[/USER]: Can you comment on [url=http://www.sciencedaily.com/releases/2015/11/151118155446.htm]this news[/url]? It looks very interesting.
Edit: [url=https://www.physicsforums.com/threads/can-gene-editing-eliminate-alzheimers-disease-comments.814472/]Missed your old insight article[/url], discussing it months ago, sorry.
“What gene are you getting inserted?”
Flying
“Saw my primary doctor for an annual checkup and even he was talking about gene editing and CRISPR!”
What gene are you getting inserted?
Saw my primary doctor for an annual checkup and even he was talking about gene editing and CRISPR!
“@Yggdrassil They’ve tried to implant CCR5delta32 HSPCs (hematopoietic stem progenitor cells) in the past, but they are swiftly outcompeted by unmodified cells – or so the paper I’ve read says (below).
[URL]http://www.pnas.org/content/111/26/9591.full[/URL]”
Although that paper makes edited HSPCs, they do not test their function [i]in vivo[/i]. The best study on this is the [url=http://www.nejm.org/doi/full/10.1056/NEJMoa1300662#t=article]NEMJ study[/url] I referenced in my previous post, which shows edited T-cells (not HSPCs) persisting at least 3.5 years after transplantation. The levels of edited T-cells do decline over time and it is not clear from the study whether there are sufficient T-cells to protect from reemergence of the virus. Transplantation of edited HSPCs is probably the strategy to pursue in the long term, though I don’t know of any data saying whether or not they will persist in patients without being outcompeted by unmodified cells (maybe someone has done a study in mice?).
@Yggdrassil They’ve tried to implant CCR5delta32 HSPCs (hematopoietic stem progenitor cells) in the past, but they are swiftly outcompeted by unmodified cells – or so the paper I’ve read says (below).
[URL]http://www.pnas.org/content/111/26/9591.full[/URL]
[USER=169968]@entropy1[/USER] You can simply design DNA sequences on computer, and literally print out chains of oligonucleotides; that’s what they do when they manufacture gene chips/DNA microarrays. On the phenome/genome thing. We know all the coding sections of the genome, we know pretty easily which proteins are made by those transcripts. The questions basically revolve around protein function (which isn’t always easy to determine), and ncRNA function (which are parts of the genome that are transcribed but do not make any proteins).
There’s a disease called Progeria which involves a single nucleotide polymorphism (and a silent one at that) in a single protein. That’s a single letter DNA change, which doesn’t even change the amino acid sequence. This is (some of) what it does to gene expression in a typical cell (by fold-change).
[IMG]http://s23.postimg.org/gtaaz42yz/progeria.jpg[/IMG]
Gene knockout by Cas9 can actually help elucidate gene function pretty well. It’s primarily an RNA-guided endonuclease, which causes a double-stranded break. When repaired via NHEJ this produces indels which will disrupt a gene or even ncRNA section of DNA, resulting a non-functional protein/product. Therefore via the effect of disrupting a gene you can try and figure out its function. And you can do whole genome function screens by designing multiple Cas9 gRNAs for the genes you want to knock out with ease and minimal outlay, which is very, very useful.
This may be a long way in the future, but on the difficulty of designing a synthetic genome, I think the click chemistry/protein catalysed capture method detailed in the article below could be used to generate an entirely new set of alternate biochemical interactions, using an entirely novel set of proteins – separate from any interactions taking place in the body. And if you know about Cas9 you’ll know about the potential for multiplexed and orthogonal transcriptional control. In terms of the development of a truly synthetic cell/biochemical computer, that would probably be the endgoal.
[URL]http://phys.org/news/2015-04-inhibitor-abnormal-protein-cancer-drugs.html[/URL]
“Roughly, what is the strategy? Oh, wait, I see you’ve linked an article [URL]http://www.nejm.org/doi/full/10.1056/NEJMoa1300662#t=article[/URL] in the blog post.
OK, I read the article quickly. It seems they take out some of the immune cells of the patient which are usually infected by HIV, make the immune cells resistant to HIV infection, then put the cells back in the patient. Here they do it just once.
In the full treatment, presumably they will have to do it multiple times, since I assume the body will generate more immune cells that are not resistant, while those that were made resistant will die off over time?”
Yes, [i]ex vivo[/i] editing of T-cell to make them resistant to HIV is the strategy. The NEMJ paper describes the T-cells persisting to at least 3.5 years after transplantation (the latest time point measured), but it is possible that the resistant cells could die off over time. The treatment could potentially be made permanent by editing bone marrow cells [i]ex vivo[/i] then transplanting in the edited bone marrow. A similar procedure was done on [url=http://www.nature.com/news/2009/090211/full/news.2009.93.html]a patient in Berlin in 2007[/url] (instead of edited bone marrow, marrow from a donor naturally carrying the CCR5 mutation was used). This patient has not seen the virus return despite remaining off of antiretroviral drugs, and is considered by many to be the [url=http://www.nbcnews.com/health/health-news/how-many-people-have-been-cured-hiv-one-n161546]only individual ever cured of HIV[/url].
“Gene editing technologies are currently being used in clinical trials to treat HIV.”
Roughly, what is the strategy? Oh, wait, I see you’ve linked an article [URL]http://www.nejm.org/doi/full/10.1056/NEJMoa1300662#t=article[/URL] in the blog post.
OK, I read the article quickly. It seems they take out some of the immune cells of the patient which are usually infected by HIV, make the immune cells resistant to HIV infection, then put the cells back in the patient. Here they do it just once.
In the full treatment, presumably they will have to do it multiple times, since I assume the body will generate more immune cells that are not resistant, while those that were made resistant will die off over time?
It is possible to design DNA sequences and make those physical DNA molecules. For example, scientists at the J. Craig Venter Institute synthesized a complete bacterial genome in 2010 and used that to create a [url=http://www2.technologyreview.com/article/423691/synthetic-cells/]synthetic cell[/url]. They currently use similar technologies to synthesize viruses for vaccine production. Instead of having virus samples sent to their lab, they can just get the virus’s DNA sequence in an email and synthesize the virus themselves. Similar work has been done to [url=http://www.nature.com/news/first-synthetic-yeast-chromosome-revealed-1.14941]synthesize yeast chromosomes[/url].
Although we are working towards having the technology to build new genomes from scratch, we lack a lot of the knowledge required to build new functions into genomes. It’s as if we have a printing press, but no one knows how to read.
This is really annoying. You do something like this, which at first glance appears to flout the policy agreements in place to not edit the human germline. You get people worked up, the researchers put something in like “it caused unintended mutations in the embryos”, and it sounds even worse. Of course it caused unintended mutations – the technology is only 2 years old. Clinical trials for HIV are presumably the ex-vivo method of generating the CCR5delta32 cells? Much less risky than in vivo editing.
I just wrote a risk assessment on CRISPR-Cas9 for therapeutic use; the tech is evolving quickly, risks of off-target effects are being mitigated every day by improvements such as tru-gRNAs, dimeric hybrid Cas9-FokI Nucleases, etc. but no way would I consider it safe to use for human therapeutics at the moment. We’ve only just had the postulation of possible methods for detecting genome wide off-target effects in the last few months – at the first instance we need to be able to detect all off-target effects, then they can be used to develop rules for enhanced target specificity. The initial computer programs (MIT, etc.) most researchers have used thus far to generate gRNAs were pretty much rendered obsolete by the new genome-wide off-target detection methods because of their inability to accurately predict off-target binding sites.
“Most of us alive today wouldn’t exist without science. Should we all fear the thing that feeds and cloths us? Maybe, but I think it’s important to put that fear in perspective. A far great horror would be a world without science. It’s important to consider the benefits along with the costs. In that light the question we should be asking ourselves in regard to genetic manipulation of humans is will the benefits outweigh the costs.”
I never said we should abandon science. But we should fear its bad consequences the same as when we fear an airplane crash. We don’t abandon airplanes, we just try to prevent crashes and still prepare for them in lots and lots of ways. I’m just saying the “airplane” of science and its “passengers” aren’t ready for the possible “crash” and haven’t been preventing it either.
I also should mention that we had several of such crashes before. Where science worked as a weapon in the wrong hands. I’m not mentioning any specific event because I don’t mean to say there is one or even several countries who should be blamed. Its something related to all of us, as humans.
“”Don’t Fear the CRISPR?”
We should fear CRISPR and that’s the only thing we can do about it. In fact I should say we should fear all sciences, because without the necessary thing I mentioned, all of it is a tool in the wrong hands and as sciences moves forward, those wrong hands can do more damage to humanity.”
Most of us alive today wouldn’t exist without science. Should we all fear the thing that feeds and cloths us? Maybe, but I think it’s important to put that fear in perspective. A far great horror would be a world without science. It’s important to consider the benefits along with the costs. In that light the question we should be asking ourselves in regard to genetic manipulation of humans is will the benefits outweigh the costs.
“Cancer’s day’s numbered? Not quite. CRISPR-Cas9 was just tried in human embryos and it had horrendous off target effects (surprise surprise).
[URL=’http://pipeline.corante.com/archives/2015/04/24/crispr_gene_editing_in_human_embryos_not_so_fast.php#comments’]http://pipeline.corante.com/archive…ing_in_human_embryos_not_so_fast.php#comments[/URL][/quote]
To be fair, the Liang et al paper did not use [URL=’http://www.sciencedirect.com/science/article/pii/S0092867413010155′]newer CRISPR methods[/URL] that can reduce off-target effects by 50-1000 fold. We’re not yet at the point where off-target effects are not a concern, but there is clearly room for improvement.
[quote]There are also other techniques that exist like TALENs and ZFNs which supposedly have lower off target effects (which come at the cost of lower efficiency), but get far less press. Don’t forget, siRNAs got tons of hype when they first came out too and were supposed to revolutionize medicine and our treatments of disease. Well, we’re still waiting for the revolution over a decade later, and many pharma companies have completely abandoned siRNAs due to their intractability for now.”
Yes, TALENs and ZFNs will probably still have uses (for example, some of the clinical trials to edit T-cell to be resistant to HIV are being done with ZFNs). A good way forward may to use CRISPR for the initial research (because it’s easier to test different targets with CRISPR), and once a good target has been identified and validated, to design TALENs or ZFNs. Of course, there are other problems such as the difficulties in cloning TALENs or the fact that the TALEN genes are quite large and difficult to package into some viral delivery vectors (like adeno-associated viruses).
I’d agree with the comparison to siRNAs, in that it will be difficult to perform gene editing in vivo. For many of the same reasons why siRNA has failed to take off as a therapy, gene editing in live humans may not ever become a worthwhile pursuit. Ex vivo gene editing to engineer immune cells or correct genetic disorders in blood cells, however, certainly seems to be a very feasible goal in the near future.
“Don’t Fear the CRISPR?”
We really shouldn’t? Let’s see!
As anything else in science, this is a tool, as much as applications are concerned, and we know that tools can be used for good or for bad! That depends on the hands holding the tool. Now its said that this tool is not ready yet because of some problems but scientists will solve these problems. The may not make it limitless, but they will push the limits away. And that makes such tools really powerful and experience teaches us that, unfortunately, its more probable for such powerful tools to be in the wrong hands. There are always people who want to get bad things out of such tools and, unfortunately, there are always scientists who are willing to serve those people.
Of course its ridiculous to say CRISPR or its innovators are the ones to be blamed. Its like blaming Einstein for the atomic bomb because without ## E=mc^2 ##, such a weapon couldn’t be built!
But then, who should be blame? What can we do?
The situation is like this. We have people who know how to pick a lock. Some of them use this skill to help people who are locked out. Some others use it to break into people’s house. You can’t blame the innovator of lock picking. The problem is with the ones who hire lock pickers for breaking into people’s house and also with the lock pickers who accept to do it. But the present situation is just a bit bigger. The tool is more powerful and so the people who hire its experts are more important people and the bad things that can come out of it has broader scope, wider range and deeper impact.
It seems to me we can do nothing about this situation. I was going to say we need to care about science ethics but it doesn’t seem to work. In fact when I think about this world and the way some people think, it seems inevitable to me that there will be some really bad consequences of this.
Don’t take me wrong. I too like to see science going forward. But my problem is, there is something as necessary as science that is being overlooked in today’s world.
So what do I think? We should fear CRISPR and that’s the only thing we can do about it. In fact I should say we should fear all sciences, because without the necessary thing I mentioned, all of it is a tool in the wrong hands and as sciences moves forward, those wrong hands can do more damage to humanity.
“Very cool development. Seems like a huge milestone. Cancer, your f days are numbered…
Since the bait was sort of “should we fear our coming ability to control our world at an entirely new level” :nb)… I’ll kick it off with, you bet! Not too much in the article convinces one (if fearful) that all the crazy things imagined in the movies couldn’t or won’t happen. The breakthrough seem to declare with seamless authority, it is now truly, more realistically possible.
Good for us! It’s still exactly what we should be trying to do, what we have always done, and what we will always do, or die tryin. What we also have, must and will do, is struggle mightily to bring the good parts of our humanity along with us. To keep making the novel human choice with what we discover we can do. What made Gattaca such a compelling story to me (a favorite) is that is one story of how that might really happen. Moon is another one. Both are nail biter’s.
I for one am totally supportive of our efforts to master what we are made of AND our worry about the risks of those efforts. :cool:
I would also love to know what the short list of anticipated breakthroughs in our burgeoning ability to control the genetic engine that creates and destroys us. Are we not living in the golden age of this field?”
Cancer’s day’s numbered? Not quite. CRISPR-Cas9 was just tried in human embryos and it had horrendous off target effects (surprise surprise).
[URL]http://pipeline.corante.com/archives/2015/04/24/crispr_gene_editing_in_human_embryos_not_so_fast.php#comments[/URL]
There are also other techniques that exist like TALENs and ZFNs which supposedly have lower off target effects (which come at the cost of lower efficiency), but get far less press. Don’t forget, siRNAs got tons of hype when they first came out too and were supposed to revolutionize medicine and our treatments of disease. Well, we’re still waiting for the revolution over a decade later, and many pharma companies have completely abandoned siRNAs due to their intractability for now.
You could fix the gene for vitamin c and end scurvy forever. Not that scurvy is much of a problem in the modern age.
“Are we not living in the golden age of this field?”
I’m sure they are just scratching the surface. Are we still in the information age or what? I think you are right on track for a “eugenics age” going straight to the genes?
Very cool development. Seems like a huge milestone. Cancer, your f days are numbered…
Since the bait was sort of “should we fear our coming ability to control our world at an entirely new level” :nb)… I’ll kick it off with, you bet! Not too much in the article convinces one (if fearful) that all the crazy things imagined in the movies couldn’t or won’t happen. The breakthrough seem to declare with seamless authority, it is now truly, more realistically possible.
Good for us! It’s still exactly what we should be trying to do, what we have always done, and what we will always do, or die tryin. What we also have, must and will do, is struggle mightily to bring the good parts of our humanity along with us. To keep making the novel human choice with what we discover we can do. What made Gattaca such a compelling story to me (a favorite) is that is one story of how that might really happen. Moon is another one. Both are nail biter’s.
I for one am totally supportive of our efforts to master what we are made of AND our worry about the risks of those efforts. :cool:
I would also love to know what the short list of anticipated breakthroughs in our burgeoning ability to control the genetic engine that creates and destroys us. Are we not living in the golden age of this field?
Comments should also appear in the article, but they are not. There are still some kinks to hammer out.
I was listening to NPR today and there was a segment on CRISPR. It’s a big deal!
“He is talking in the third person now? ;)
If this is an automatic post, I wonder how it got into the right forum.
Great post!”
Yup, this was an automatic post that’ll pop up whenever I post to the Insights blog. Greg has done a great job getting the blog integrated with the forums so that things like this are possible.
Glad you liked the post!
”
If this is an automatic post, I wonder how it got into the right forum.
”
When you write the blog entry, you select which forum is relevant.
This thread is created so people can comment on the blog.
He is talking in the third person now? ;)
If this is an automatic post, I wonder how it got into the right forum.
Great post!
Greg, just a technical comment. When the graphics in this article are under the mouse, they get dim and a blue point with two arrows appears. Clicking on that arrow, the graphics get larger. But as the background of the graphics is transparent, it becomes barely visible.
gg
Would it be possible, in the future, to -compose- DNA strands that are designed on a computer? You’ll have to do statistical phenotype-genome calculations then, I imagine.
Just how far away are we from using this against cancer? Next 5 years or 10?
Gene editing technologies are currently being used in clinical trials to treat HIV. There are potential applications in cancer, but none that I am aware of that are close to being close to clinical trials. We’re probably at least a decade away.
Here is the story I heard on NPR, you can listen here too http://www.npr.org/blogs/health/2015/04/23/401655818/critics-lash-out-at-chinese-scientists-who-edited-dna-in-human-embryos