Posterior Predictive Distributions in Bayesian Statistics
Table of Contents
Confessions of a moderate Bayesian, part 4
Bayesian statistics by and for non-statisticians
Read part 1: How to Get Started with Bayesian Statistics
Read part 2: Frequentist Probability vs Bayesian Probability
Read part 3: How Bayesian Inference Works in the Context of Science
Predictive distributions
A predictive distribution is a distribution that we expect for future observations. In other words, instead of describing the mean or the standard deviation of the data we already have, it describes unobserved data.
Predictive distributions are not unique to Bayesian statistics, but one huge advantage of Bayesian statistics is that the Bayesian posterior predictive distribution considers the uncertainty of the parameters. This makes the Bayesian posterior predictive distribution a better representation of our best understanding of the process that generated the data.
Basic concept
One of the core ideas of statistics is to make a model of the data-generating process. Usually, this model has some unknown parameters that are estimated from the data itself. Often those parameters tell us something important about the system, so often we are just interested in the parameters. But, once we have those parameters, we can turn around and use the model with the parameters to predict what we would expect as new observations.
In Bayesian statistics, we don’t end up with point estimates of the parameters but samples of the posterior joint distribution of the parameters. To get samples of the posterior predictive distribution we take a sample from the posterior parameter distribution, get a set of parameters, plug those parameters into the model, and generate a predicted sample of the data. The distribution of those predictions is the posterior predictive distribution.
Uses
The main use of the posterior predictive distribution is to check if the model is a reasonable model for the data. We do this by essentially simulating multiple replications of the entire experiment. For each data point in our data, we take all the independent variables, take a sample of the posterior parameter distribution, and use those to generate a sample of the posterior predictive distribution of the data. We can repeat this multiple times, generating multiple complete replications of the same experiment. If our simulated data looks like our actual data, then the model is reasonable.
Another use of the posterior predictive distribution is to see what we would expect to change because of some proposed intervention. In this case, we follow a process like what we described above, except that instead of using the independent variables as specified in the actual experiment we modify the independent variables to reflect our proposed intervention. Then we can simulate what difference that change would make compared to the original data or compared to a different intervention.
Checking model quality
First, I will show how to use the posterior predictive distribution to check and refine the model.
Chicken growth models
In a previous Insights I used the “chickweights” data set. I now return to that data set. This data set examines the growth (weight) of a large number of baby chickens from hatching to 21 days. The chicks were split into four groups which were fed different diets.
Generating the posterior predictive distribution
First, we load some necessary libraries and the data itself. And as before we will exclude the data from less than one week of age.
library(rstanarm)
library(ggplot2)
library(bayesplot)
library(emmeans)
data("ChickWeight")
dat <- subset(ChickWeight,Time > 7)Next, we will specify our model and sample the posterior distribution of the model parameters. In this case, we will model the weight of each chick as being normally distributed with a mean that depends linearly on the age of the chick (Time) and on the feed being used (Diet) for that chick.
set.seed(4567) post <- stan_lm( weight ~ Diet*Time, data = dat, prior = R2(.7))
Once we have the posterior distribution of the model parameters then we can generate the posterior predictive distribution of the data. In this case, we will generate 500 predicted replicates of the original experiment. We will then plot the probability density function (PDF). We will display the PDF of the original data together with the PDF of the first 50 predicted replicates to see how well they match.
w <- dat$weight wrep <- posterior_predict(post,draws = 500) ppc_dens_overlay(w, wrep[1:50,])
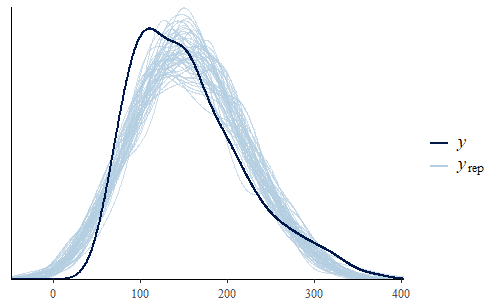
As we can see, the posterior predicted distribution does not match the actual data very well. So let’s dig a little deeper and see if we can find the cause of this discrepancy.
Examining model discrepancies
First, we will do a scatter plot of the average of the posterior predicted data vs the original data. This will help us look for patterns of deviations in our predictions vs the data.
ppc_scatter_avg(w, wrep)
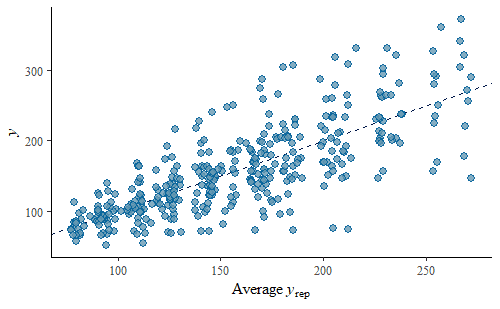
The simulated data looks pretty good overall, there is no systematic departure from the identity line. However, we do notice that the variance is substantially smaller for lower weights than for higher weights. We can explore this further by calculating some summary statistics on the predicted replicates and compare that to the same summary statistic calculated on the original data. We will calculate the mean and the min of the data and each replicate.
ppc_stat(w, wrep, stat = "mean") ppc_stat(w, wrep, stat = "min")
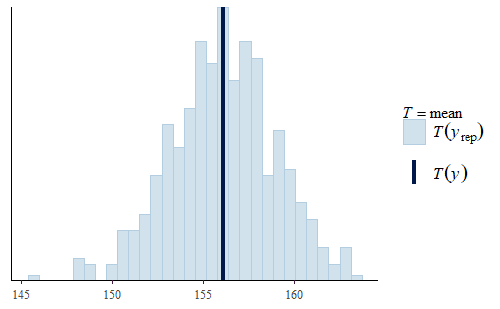
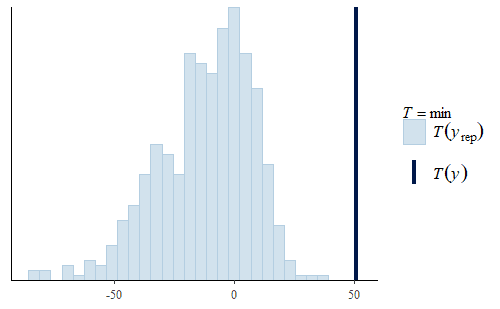
Here, we see two interesting facts.
First, the mean is quite good. The actual mean of the original data is very representative of the means of the replicates. This is quite common since the mean was specifically modeled. So the model will accurately represent the mean of the data even though the model does not capture other features of the data. If all you wanted to do was make inferences about the mean, then you could use this model as it is. This is an important point, a model is not just “good” or “bad”, but the value of a model depends on what it will be used for.
Second, the min is not very good. The actual min of the original data is well above the range of the mins of the replicates. The minimum weight in the original data is about 50 g, but the model usually has negative minimum weights. Not only is that a strong discrepancy with the data, but negative weights don’t make any physical sense either.
Improving the model
Luckily, we can make one single change that will address both the increasing variance with larger predictions and also the negative weights. Instead of modeling the weights themselves as normally distributed, we will model the log of the weights as normally distributed.
post <- stan_lm( log(weight) ~ Diet * Time, data = dat, prior = R2(.7)) w <- dat$weight wrep <- exp(posterior_predict(post,draws = 500)) ppc_dens_overlay(w, wrep[1:50,]) ppc_stat(w, wrep, stat = "min")
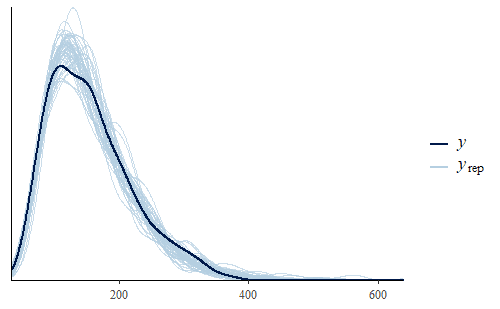
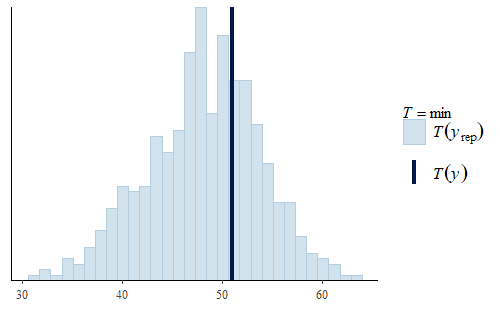
This time we see that the PDF of the posterior predictive distribution closely matches the PDF of the original data. Furthermore, the min is much more reasonable, with a smaller range that includes no negative weights and the actual data min is very representative of the mins of the replicates.
Using the model for custom information
At this point, we now feel confident in using our model to make predictions about the min as well as the mean. But having found a model which seems to capture the data faithfully we can now turn to look at features that might be of more direct interest. For example, suppose that our customers purchase chicks by weight at 21 days, but will not accept any chicks less than 150 g. Then our main interest might be the total weight (in kg) of all birds over 150 g at day 21 which would be (proportional to) the value of the chicks.
chickvalue <- function(x){
return(sum((dat$Time == 21)*(x>150)*(x)/1000))
}
ppc_stat(w, wrep, stat = "chickvalue")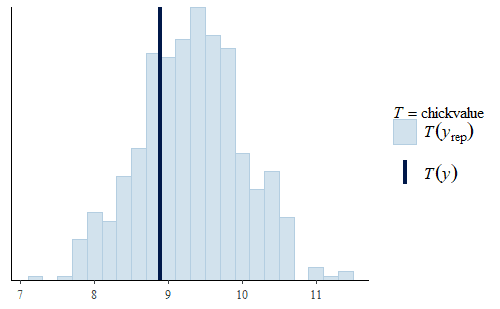
Here we see that even though the model was not specifically fit to the “chickvalue” measure, because it is a faithful representation of the data overall, the model seems to be suitable to represent this aspect of the data as well.
Note that the model still has limitations. For example, even though the data contains information about the number of chicks that died during the 21 days, we did not model that. So if we look at our replicate data sets we will find that they all have the same rate of chick survival
livechick <- function(x){
return(sum(dat$Time == 21)/sum(dat$Time == 8))
}
ppc_stat(w, wrep, stat = "livechick", binwidth = .01) + coord_cartesian(xlim = c(.70, 1))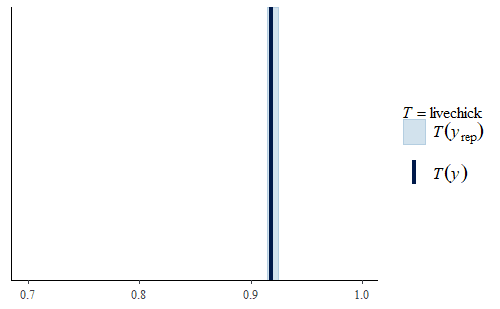
Heteroskedastic data
Shifting gears, I will now discuss a data set that came up in some research at work recently. It consists of six different measurement techniques (all measuring the same quantity) on several samples of standards with known values of the measured quantity. Because the samples have known values we could determine the error of each measurement. We were attempting to characterize the bias and precision of the six techniques.
After playing around with different models for more than a full day at work the best model that I could find was the following
set.seed(4567)
post <- stan_lmer(Err ~ Correction|Segment, data = dat.pdff,
diagnostic_file = file.path(tempdir(), "post.csv"), contrasts = FALSE, adapt_delta = 0.999)
y <- dat.pdff$Err
yrep <- posterior_predict(post,draws = 500)
ppc_dens_overlay(y, yrep[1:50,])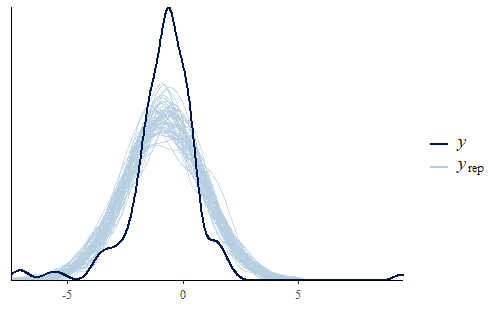
As you can see, the PDF of the replicates is different from the PDF of the original data. We see that the interquartile range of the model is too large and yet the model’s min and max are both not extreme enough. The model has a fairly standard Gaussian shape while the data has a sharp peak and heavy tails.
Upon further investigation, it became clear that the issue was some strong heteroskedacity of the data. Specifically, although all the methods seemed to have a similar small bias, two of the methods were substantially less precise than the others.
ggplot(dat.pdff,aes(x=Correction,y=Err)) + geom_boxplot(outlier.shape = NA) + geom_jitter(aes(color = Segment), alpha = 0.5)
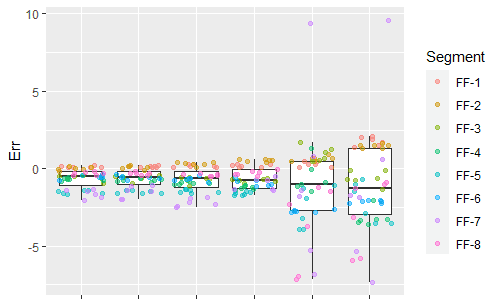
This is an issue that comes up frequently in real data where the variances are not equal. However, most statistical methods assume that the variance is equal across the data. This is such a standard assumption that many researchers forget about it entirely. However, Bayesian analysis can go beyond this limitation and we can directly model the heteroskedasity. So I ran a new analysis, this time modeling the errors as being normally distributed with a mean and a standard deviation that both depend on the measurement technique.
post <- brm(bf(Err ~ Correction, sigma ~ Correction), data = dat.pdff, family = gaussian()) y <- dat.pdff$Err yrep <- posterior_predict(post,draws = 500) ppc_dens_overlay(y, yrep[1:50,]) ppc_stat(y, yrep, stat = "median") ppc_stat(y, yrep, stat = "min") ppc_stat(y, yrep, stat = "max")
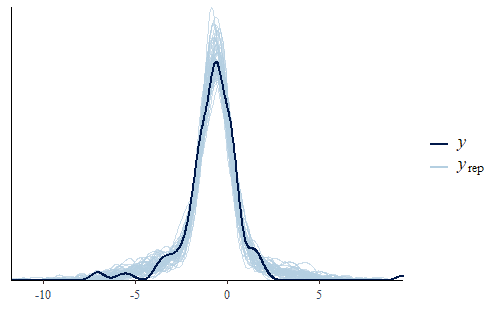
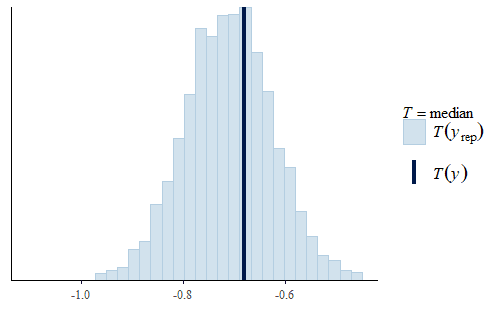
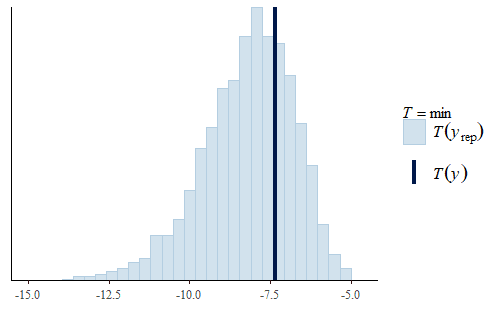
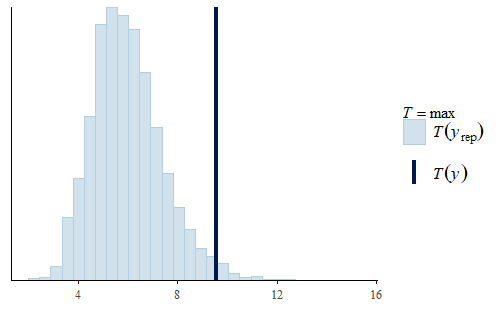
This time the PDF of the actual data is representative of the PDF of the posterior predicted replicates. The IQR, min, and max are also all reasonably well represented.
Interestingly, the max of the data is somewhat on the high side of the range of maxes of the replicates. The max of the data is a single data point that appears to be a bit of an outlier. I do not reject data simply because it seems like an outlier, and since I did not have any experimental reason for rejecting that data point we retained it in the analysis.
I did not do any manual “tweaking” or other games to reduce the impact of that data point. But the Bayesian method itself automatically included a bit of healthy skepticism about that data point. It chose a model where that point is not implausible, but it also chose a model where we do not expect quite such an extreme outlier in most of our predicted replicates. If I were going to do some manual “tweaking” of the model, that is exactly what I would have chosen: a model that is informed by the possible outlier but not entirely beholden to it.
Investigating treatment predictions
By far, most of my experience with posterior predictive distributions has centered around the use case of critiquing and refining the model. However, the posterior predictive distribution is so named because you can use it to make predictions. Instead of making a prediction based on the actual experiment, you can make predictions based on hypothetical data using the model developed from the actual data.
For example, let’s return to the ChickWeights data. Now we have a good model that looks like it plausibly captures the main features of the data. In particular, we think that the model does a good job representing how much chicken we expect to be able to sell (total weight of all chicks larger than 150 g at 21 days). So now, let’s do two simulations, one where we simulate feeding all the chicks with diet 3 and another where we simulate feeding all the chicks with diet 1. To generate the data for simulation we do the following:
dat.diet3 <- dat dat.diet3$Diet[] <- 3 dat.diet1 <- dat dat.diet1$Diet[] <- 1
Then we generate and plot the posterior predictions using the following code:
w <- dat$weight wrep <- exp(posterior_predict(post, draws = 500, newdata = dat.diet1)) ppc_stat(w, wrep, stat = "chickvalue", binwidth = 1) wrep <- exp(posterior_predict(post, draws = 500, newdata = dat.diet3)) ppc_stat(w, wrep, stat = "chickvalue", binwidth = 1)
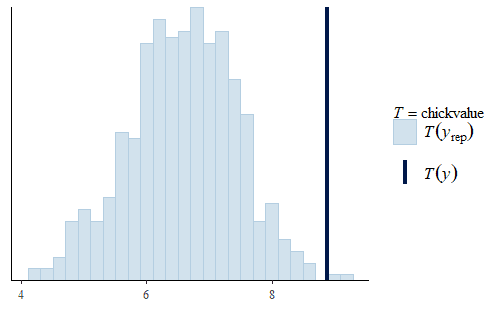
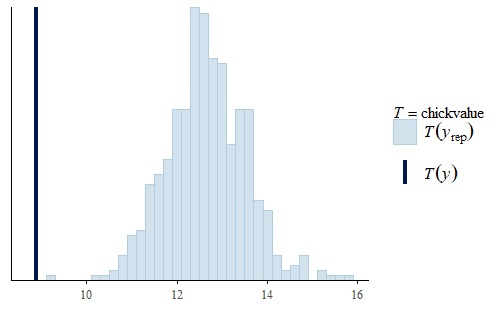
So based on this posterior prediction, we would expect that if we were to use exclusively Diet 1 then we would get around 7 kg of sellable birds for this size batch. In contrast, for Diet 3 we would expect around 13 kg of sellable birds. That is a substantial difference. Based on this model and the experiment, we could compare the costs of raising the chicks to 21 days using the two different diets and thus determine which is the more profitable approach. And note that this approach completely avoids any inappropriate overemphasis on whether the difference is “statistically significant”.
Summary
I am still new to using posterior predictive distributions, but they have already become my favorite tool for examining, criticizing, and improving a model. Using arbitrary statistics that are individually tailored to a specific need is intriguing to me. I especially like this tool for checking the appropriateness of the model for any statistic.
I also have found that sometimes model parameters themselves can become difficult to interpret. I have occasionally avoided a specific model, not because I had any valid statistical or experimental reason to do so, but because the parameters were too confusing to interpret. The posterior predictive distribution can help with that by focusing attention on the predicted hypothetical observations, especially since the observations usually represent a quantity that is of direct interest to the researchers. Overall, I have found the posterior predictive distribution to be an exceptionally useful and flexible tool.
Education: PhD in biomedical engineering and MBA
Interests: family, church, farming, martial arts



It would be interesting to define what "best prediction" means in this case. The various senses of "best" for point estimators are well know ( unbiased, minimum variance, maximum liklihood, etc.). But what does it mean for an estimator whose outcome is a distribution to be the best estimator for a distribution?
"I certainly cannot be rigorous, but I think that I adequately demonstrated several very useful features of the posterior predictive distribution. In particular, one feature that I would like from a “best estimator” distribution is that it neither ignore outliers nor overfit them. I was quite excited to find exactly that in my experience applying this method to real-world data. It was one of those things that I didn’t know I wanted until I saw it.
I disagree. Even if nature has some underlying process with a single value of the parameter, we don't know what the value of that parameter is. So the posterior predictive distribution is the best prediction we can make of future observations, given our current data.
"
It would be interesting to define what "best prediction" means in this case. The various senses of "best" for point estimators are well know ( unbiased, minimum variance, maximum liklihood, etc.). But what does it mean for an estimator whose outcome is a distribution to be the best estimator for a distribution?
That said, that section is not intended to be even an ambiguous definition of the posterior predictive distribution. It is simply a description of how you can obtain samples of the posterior predictive distribution from samples of the posterior distribution.
"we generate batches of data by repeating the following process M times: Generate one value θi of Θ from the distribution g. Then generate a lot of values xi,j,j=1,2,3,…N of X from the distribution F(x,θi)."Typically ##N=1##. In principle you could have ##N>1## but that is not generally done. That is why I said "generate a predicted sample". I don't know if there is a specific reason for that, but it is what I have always seen done in the literature and so I have copied that.
"probability model for Y does not match the Bayesian assumption we adopted because the Bayesian assumption is that a single value of the parameter Θ was used when Nature generated the data D."I disagree. Even if nature has some underlying process with a single value of the parameter, we don't know what the value of that parameter is. So the posterior predictive distribution is the best prediction we can make of future observations, given our current data. Any single value of the parameters that we select would underestimate our uncertainty in the prediction. Accounting for our uncertainty in the parameters is definitely Bayesian, selecting a single value of the parameters would not be a good Bayesian approach even if we believe that nature has such a single value.
"Each simulation of a batch of data from a distribution F(x,θi) can be used to make point estimates of a different parameter γi of the distribution F(x,θi). ( For example, if θi is the (population) mean of the distribution then the sample variance of the data we generated can be used make a point estimate of the variance σi2 of F(x,θi).) The simulation process provides different batches of data, so it provides a histogram of γi,i=1,2,…M that estimates the posterior distribution of γ. We can make a point estimate of γ using the original data D and see where this estimate falls on the histogram of the simulated data for γ."I haven't seen it done that way, but I don't know why it couldn't be done that way. Each individual batch would systematically understate the uncertainty in the predicted data, but I am not sure that would mean that the resulting histogram of ##\gamma## would similarly have artificially reduced uncertainty. It would be plausible to me that the inter-batch variation would adequately represent our uncertainty in ##\gamma##.
To get samples of the posterior predictive distribution we take a sample from the posterior parameter distribution, get a set of parameters, plug those parameters into the model, and generate a predicted sample of the data. The distribution of those predictions is the posterior predictive distribution.
Source https://www.physicsforums.com/insights/posterior-predictive-distributions-in-bayesian-statistics/
"
That definition of the posterior predictive distribution is ambiguous – perhaps the intent is use examples to make it precise, but I'd have to learn the computer code to figure it out.
Suppose we have a random variable ##X## whose distribution is given by a function ##F(x,\Theta)## where ##\Theta## is the parameter of ##F##. We make the Bayesian assumption that ##\Theta## has a prior distribution. Based on a set of data ##D## we get a posterior distribution ##g(\theta)## for ##\Theta##. Following the definition given above, we generate batches of data by repeating the following process ##M## times: Generate one value ##\theta_i## of ##\Theta## from the distribution ##g##. Then generate a lot of values ##x_{i,j}, j = 1,2,3,…N## of ##X## from the distribution ##F(x,\theta_i)##.
The result of this simulation process is a "distribution of distributions". If we lump all the ##X_{i,j}, i = 1,2,..M, j = 1,2,…N## data into one histogram and consider it an approximation for the distribution of a single random variable ##Y## then the probability model for ##Y## does not match the Bayesian assumption we adopted because the Bayesian assumption is that a single value of the parameter ##\Theta## was used when Nature generated the data ##D##.
( Of course, I'm speaking as an "objective" Bayesian. If one thinks of prior and posterior distributions as measuring "degrees of belief" then who is to say whether lumping the simulation data is into a single histogram is valid?)
It seems to me that the complete definition of a "predictive distribution" should say that it is a posterior distribution of some parameter, which may be different than the parameter that was assigned a prior distribution.
Each simulation of a batch of data from a distribution ##F(x,\theta_i)## can be used to make point estimates of a different parameter ##\gamma_i## of the distribution ##F(x,\theta_i)##. ( For example, if ##\theta_i## is the (population) mean of the distribution then the sample variance of the data we generated can be used make a point estimate of the variance ##\sigma^2_i## of ##F(x,\theta_i)##.) The simulation process provides different batches of data, so it provides a histogram of ##\gamma_i, i = 1,2,…M## that estimates the posterior distribution of ##\gamma##. We can make a point estimate of ##\gamma## using the original data ##D## and see where this estimate falls on the histogram of the simulated data for ##\gamma##.
I hope that's right.
"Yes, that formula is indeed correct and it looks like you did the arithmetic correct also. What you are seeing is the correct and expected result.
"
Problem: how can p(d/-) be less than 0.001?
"Remember that p(d) was 0.001 is our prior belief. So without even having the test we were pretty convinced that he didn’t have the disease simply because the disease is rare. When we get a negative result for the test we are going to be even more certain that he doesn’t have the disease. So a negative test will always reduce our belief that the disease is present.
It wouldn't make sense to say "Yesterday I was pretty confident that he is not sick since it is a rare disease, but today after the test was negative I am concerned that he is probably sick". Instead what we would say is "Yesterday I was pretty confident that he is not sick since it is a rare disease, and today after the test was negative I am utterly convinced that he is not sick."
As @stevendaryl mentioned believing that he is sick requires two unlikely things, first he had to be the poor rare person that actually has the disease, and second he has to coincidentally be the rare sick person that gets a false negative. Believing that he is well requires two likely things, first he had to be in the bulk of the population that is well and second he has to be in the bulk of the well population that gets a true negative. Two likely things are much more likely than two unlikely things. So the direction of change, to less than 0.001 is correct.
"
And by 3 orders of magnitude???
"It is actually about 2 orders of magnitude. From ##1 \ 10^{-3}## to ##1 \ 10^{-5}##. As a rough estimate you can look at the Bayes factor for a negative test. The Bayes factor is $$\frac{P(-|d)}{P(-|d*)}=\frac{1-0.99}{0.99}\approx 0.01$$ so we do expect it to change by about two orders of magnitude.
You did the math correctly.
Problem: how can p(d/-) be less than 0.001? And by 3 orders of magnitude???
Get me past this hurdle so I can start reading your 1st blog!
Thx.
BTW I did p(d/+) and it came out right at about 9%.
"
Well, p(d/-) is roughly the probability of two unlikely things happening: That he has the disease, and that the test is wrong.
However, to whet my appetite, help me if you care to re the following:
I have a population of 1000. I wantr to test an individual for a certain disease d.
Known: average no. of actual diseased individuals = 1 in 1000. My test is 99 effective in that there is only a 1 chance in 100 that a diseased ind. will register a negative test result. My test is also 99% effective in that it reports a healthy person with a negative test result 99% of the time.
Calling d = diseased, d* = healthy, + a positive test result and – a negative result, the post-test result should be
## p(d/-) = \frac {p(-/d) p(d)} {p(-/d) p(d) + p(d*) p(-/d*)}. ##
I hope that's right.
Now, putting numbers in it, the pre-test numbers are (I think)
## p(-/d) = 0.01 ##
## p(d) = 0.001 ##
## p(d*) = 0.999 ##
## p(-/d*) = 0.99 ##
giving ## p(d/-) = \frac { .01 x .001} {.01 x .001 + .999 x .99} ~ 10^{-5}##.
Problem: how can p(d/-) be less than 0.001? And by 3 orders of magnitude???
Get me past this hurdle so I can start reading your 1st blog!
Thx.
BTW I did p(d/+) and it came out right at about 9%.