- #1
fab13
- 318
- 6
Hello,
I follow the post https://www.physicsforums.com/threads/cross-correlations-what-size-to-select-for-the-matrix.967222/#post-6141227 .
It talks about the constraints on cosmological parameters and forecast on futur Dark energy surveys with Fisher's matrix formalism.
Below a capture of Martin White's paper : this is the part of building the Fisher matrix with eq(17) where C_xy is the covariance matrix of observables.
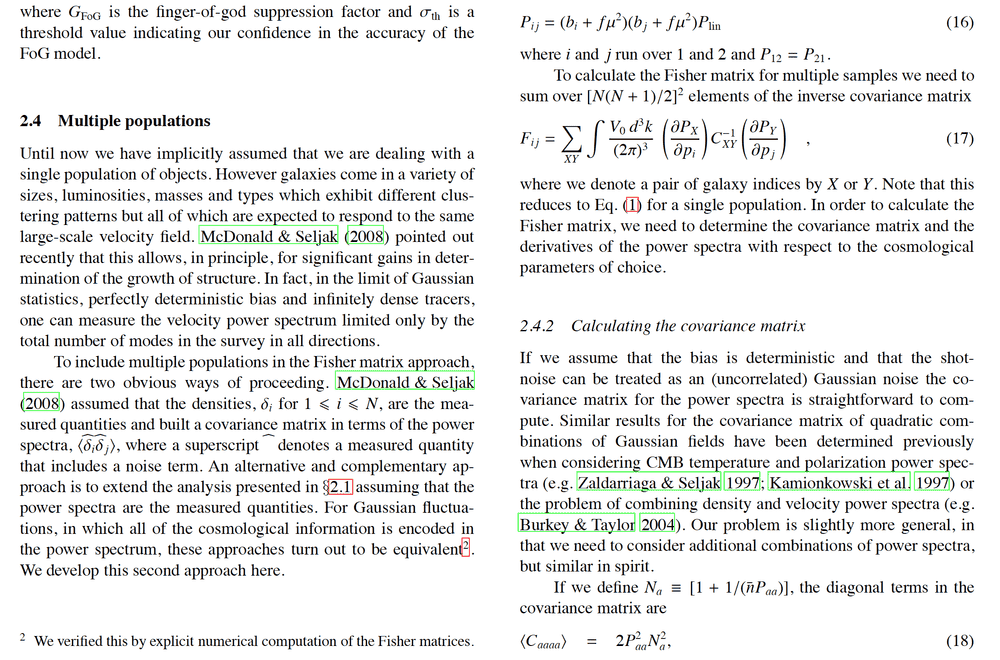
Actually, I try to do cross-correlations with multi-populations (in my case, I have 3 populations of galaxies : BGS, LRG, ELG).
My main problem is about the formula : ##N_{a} = \bigg[1+\dfrac{1}{(\bar{n}\,P_{aa})}\bigg]## : indeed, some of my redshift bins have a ##\bar{n}## equal to 0 and that makes diverge the definition of ##N_{a}##.
Here is the bias (first column = redshift, (2,3,4) columns = bias_{BGS}, bias_{LRG}, bias_{ELG} and (5,6,7) columns = n_{BGS}, n_{LRG}, n_{ELG} :
# z b1 b2 b3 n1 n2 n3 f(z) G(z)
1.7500000000e-01 1.1133849956e+00 0.0000000000e+00 0.0000000000e+00 2.8623766865e+03 0.0000000000e+00 0.0000000000e+00 6.3004698923e-01 6.1334138190e-01
4.2500000000e-01 1.7983127401e+00 0.0000000000e+00 0.0000000000e+00 2.0412110815e+02 0.0000000000e+00 0.0000000000e+00 7.3889083367e-01 6.1334138190e-01
6.5000000000e-01 0.0000000000e+00 1.4469899900e+00 7.1498329000e-01 0.0000000000e+00 8.3200000000e+02 3.0900000000e+02 8.0866367407e-01 6.1334138190e-01
8.5000000000e-01 0.0000000000e+00 1.4194157200e+00 7.0135835000e-01 0.0000000000e+00 6.6200000000e+02 1.9230000000e+03 8.5346288717e-01 6.1334138190e-01
1.0500000000e+00 0.0000000000e+00 1.4006739400e+00 6.9209771000e-01 0.0000000000e+00 5.1000000000e+01 1.4410000000e+03 8.8639317400e-01 6.1334138190e-01
1.2500000000e+00 0.0000000000e+00 0.0000000000e+00 6.8562140000e-01 0.0000000000e+00 0.0000000000e+00 1.3370000000e+03 9.1074033422e-01 6.1334138190e-01
1.4500000000e+00 0.0000000000e+00 0.0000000000e+00 6.8097541000e-01 0.0000000000e+00 0.0000000000e+00 4.6600000000e+02 9.2892482272e-01 6.1334138190e-01
1.6500000000e+00 0.0000000000e+00 0.0000000000e+00 6.7756594000e-01 0.0000000000e+00 0.0000000000e+00 1.2600000000e+02 9.4267299703e-01 6.1334138190e-01
As you can see, there is an overlapping between b2 and b3, like n2 and n3 (3 redshifts bins that are overlapped).
Question 1) How can I deal with the values ##b_{i}## and ##n_{j}## equal to zero with the definition of ##N_{a} = \bigg[1+\dfrac{1}{(\bar{n}\,P_{aa})}\bigg]## ?
I mean how to avoid a division by zero while calculating the different coefficients ##C_{ijkl}## of covariance matrix ?
Question 2) Into my code, can I set a high value for ##N_{a}## when my redshift bin has a bias or n(i) equal to zero ?
Question 3) Do you think in paper that ##C_{XY}## is a 3x3 or 4x4 matrix ? a colleague told me it could be a 3x3 matrix whereas in my previous post ( https://www.physicsforums.com/threa...to-select-for-the-matrix.967222/#post-6141227 ), it seems to be considered as a 4x4 structure.
Regards
I follow the post https://www.physicsforums.com/threads/cross-correlations-what-size-to-select-for-the-matrix.967222/#post-6141227 .
It talks about the constraints on cosmological parameters and forecast on futur Dark energy surveys with Fisher's matrix formalism.
Below a capture of Martin White's paper : this is the part of building the Fisher matrix with eq(17) where C_xy is the covariance matrix of observables.
Actually, I try to do cross-correlations with multi-populations (in my case, I have 3 populations of galaxies : BGS, LRG, ELG).
My main problem is about the formula : ##N_{a} = \bigg[1+\dfrac{1}{(\bar{n}\,P_{aa})}\bigg]## : indeed, some of my redshift bins have a ##\bar{n}## equal to 0 and that makes diverge the definition of ##N_{a}##.
Here is the bias (first column = redshift, (2,3,4) columns = bias_{BGS}, bias_{LRG}, bias_{ELG} and (5,6,7) columns = n_{BGS}, n_{LRG}, n_{ELG} :
# z b1 b2 b3 n1 n2 n3 f(z) G(z)
1.7500000000e-01 1.1133849956e+00 0.0000000000e+00 0.0000000000e+00 2.8623766865e+03 0.0000000000e+00 0.0000000000e+00 6.3004698923e-01 6.1334138190e-01
4.2500000000e-01 1.7983127401e+00 0.0000000000e+00 0.0000000000e+00 2.0412110815e+02 0.0000000000e+00 0.0000000000e+00 7.3889083367e-01 6.1334138190e-01
6.5000000000e-01 0.0000000000e+00 1.4469899900e+00 7.1498329000e-01 0.0000000000e+00 8.3200000000e+02 3.0900000000e+02 8.0866367407e-01 6.1334138190e-01
8.5000000000e-01 0.0000000000e+00 1.4194157200e+00 7.0135835000e-01 0.0000000000e+00 6.6200000000e+02 1.9230000000e+03 8.5346288717e-01 6.1334138190e-01
1.0500000000e+00 0.0000000000e+00 1.4006739400e+00 6.9209771000e-01 0.0000000000e+00 5.1000000000e+01 1.4410000000e+03 8.8639317400e-01 6.1334138190e-01
1.2500000000e+00 0.0000000000e+00 0.0000000000e+00 6.8562140000e-01 0.0000000000e+00 0.0000000000e+00 1.3370000000e+03 9.1074033422e-01 6.1334138190e-01
1.4500000000e+00 0.0000000000e+00 0.0000000000e+00 6.8097541000e-01 0.0000000000e+00 0.0000000000e+00 4.6600000000e+02 9.2892482272e-01 6.1334138190e-01
1.6500000000e+00 0.0000000000e+00 0.0000000000e+00 6.7756594000e-01 0.0000000000e+00 0.0000000000e+00 1.2600000000e+02 9.4267299703e-01 6.1334138190e-01
As you can see, there is an overlapping between b2 and b3, like n2 and n3 (3 redshifts bins that are overlapped).
Question 1) How can I deal with the values ##b_{i}## and ##n_{j}## equal to zero with the definition of ##N_{a} = \bigg[1+\dfrac{1}{(\bar{n}\,P_{aa})}\bigg]## ?
I mean how to avoid a division by zero while calculating the different coefficients ##C_{ijkl}## of covariance matrix ?
Question 2) Into my code, can I set a high value for ##N_{a}## when my redshift bin has a bias or n(i) equal to zero ?
Question 3) Do you think in paper that ##C_{XY}## is a 3x3 or 4x4 matrix ? a colleague told me it could be a 3x3 matrix whereas in my previous post ( https://www.physicsforums.com/threa...to-select-for-the-matrix.967222/#post-6141227 ), it seems to be considered as a 4x4 structure.
Regards
Attachments
Last edited:


