Sneaky6666
- 14
- 0
I have a unique problem and looking for a known fast algorithm for it. I have an unweighted but directed graph `G`. I then have a subset collection of nodes `S` existing in `G`. I want to find the minimum sub tree in `G` such that it contains all the nodes in `S`.
I so far found Chu-Liu/Edmonds algorithm but it is for finding the MST for all nodes in the graph.
Does anyone know how to do this for a subset of nodes?
Example
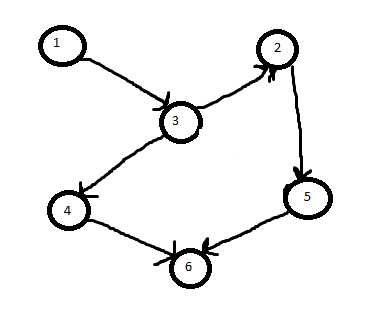
If the picture is G. And S is the node set {1, 6}. Then the solution (smallest sub tree) is the subset graph of {1, 3, 4, 6} since it includes the nodes of S. It wouldn't be {1, 3, 2, 5, 6} (even though it includes the nodes of S) because that is larger.
Thanks
I so far found Chu-Liu/Edmonds algorithm but it is for finding the MST for all nodes in the graph.
Does anyone know how to do this for a subset of nodes?
Example
If the picture is G. And S is the node set {1, 6}. Then the solution (smallest sub tree) is the subset graph of {1, 3, 4, 6} since it includes the nodes of S. It wouldn't be {1, 3, 2, 5, 6} (even though it includes the nodes of S) because that is larger.
Thanks
Last edited: