Malamala
- 345
- 28
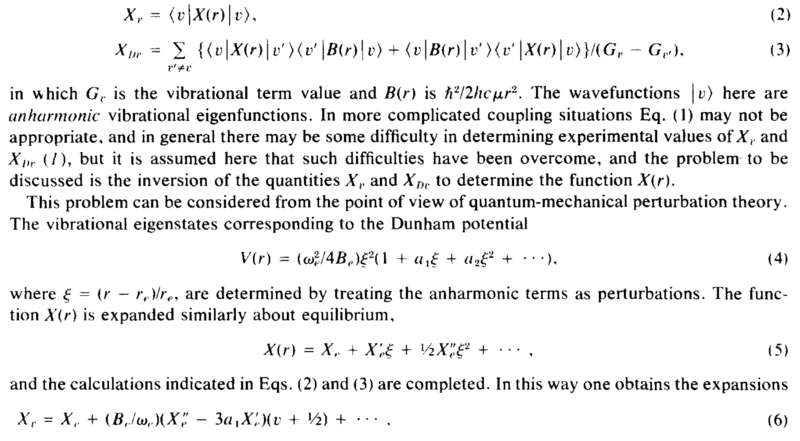
Hello! I am confused about the derivation in the screenshot below. This is in the context of a diatomic molecular potential, but the question is quite general. Say that the potential describing the interaction between 2 masses, as a function of the radius between them is given by the anharmonic oscillator potential in eq 4., where ##r_e## is the equilibrium separation. What I need is to calculate the expectation value of a new variable, ##X(r)## in between 2 wavefunctions of such a potential, eq. 2 (please ignore eq. 3 and most of the comments in the paragraph after, as they are not related to my question). They Taylor expand ##X(r)## as in eq. 5 and then they claim that from there it follows that ##X_\nu## (eq. 2) is given by eq. 6. Can someone help me understand how to go from eq. 2, 4 and 5 to eq. 6? Thank you!