- #1
- 3,759
- 4,199
Published this week in the journal Science, researchers report that they have devised a eight letter alphabet for DNA and RNA:
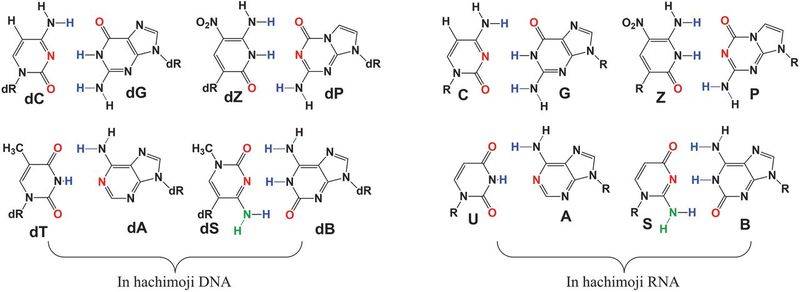
The work builds off of previous work, which had expanded the genetic alphabet to six letters. The researchers call their eight-lettered nucleic acids "hachimoji," Japanese for eight letters. The four new letters seem to function just as well as the original four DNA letters, and the researchers were even able to enzymatically copy the eight-lettered DNA molecules into eight-lettered strands of RNA.
Hoshika et al. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 363: 884 (2019)
http://science.sciencemag.org/content/363/6429/884
Abstract:
Popular press summary: https://www.nytimes.com/2019/02/21/science/dna-hachimoji-genetic-alphabet.html
The work builds off of previous work, which had expanded the genetic alphabet to six letters. The researchers call their eight-lettered nucleic acids "hachimoji," Japanese for eight letters. The four new letters seem to function just as well as the original four DNA letters, and the researchers were even able to enzymatically copy the eight-lettered DNA molecules into eight-lettered strands of RNA.
Hoshika et al. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 363: 884 (2019)
http://science.sciencemag.org/content/363/6429/884
Abstract:
.We report DNA- and RNA-like systems built from eight nucleotide “letters” (hence the name “hachimoji”) that form four orthogonal pairs. These synthetic systems meet the structural requirements needed to support Darwinian evolution, including a polyelectrolyte backbone, predictable thermodynamic stability, and stereoregular building blocks that fit a Schrödinger aperiodic crystal. Measured thermodynamic parameters predict the stability of hachimoji duplexes, allowing hachimoji DNA to increase the information density of natural terran DNA. Three crystal structures show that the synthetic building blocks do not perturb the aperiodic crystal seen in the DNA double helix. Hachimoji DNA was then transcribed to give hachimoji RNA in the form of a functioning fluorescent hachimoji aptamer. These results expand the scope of molecular structures that might support life, including life throughout the cosmos.
Popular press summary: https://www.nytimes.com/2019/02/21/science/dna-hachimoji-genetic-alphabet.html
