- #1
paunonen89
- 4
- 0
Hey, I'm looking for some help understanding how to solve these equations. The math is just a little bit beyond me. I would like to plot the equations over time to see how they are behaving.

These refer to the following equations:
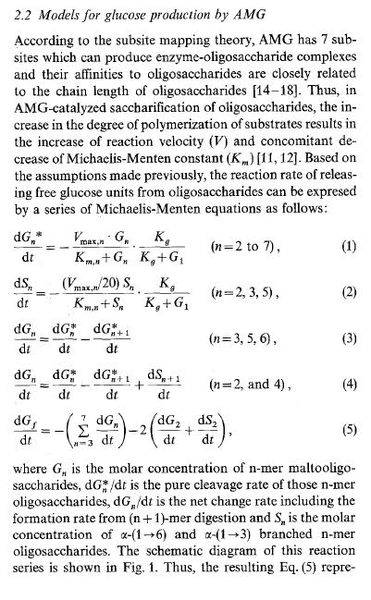
Do I just sub all of these equations into number 14 to make a giant one to solve?
I have all the constants, the unknown values are G1-G7 and S2,S3,S5. Also given are initial values for Gn/S0, Sn/S0 and Vmax,n
The ultimate goal of this is to make a computer simulation, coded in vb.net. If you could also suggest a numerical integragtion method I can possibly turn it into a vb.net program that can solve the system...Thanks!

These refer to the following equations:
Do I just sub all of these equations into number 14 to make a giant one to solve?
I have all the constants, the unknown values are G1-G7 and S2,S3,S5. Also given are initial values for Gn/S0, Sn/S0 and Vmax,n
The ultimate goal of this is to make a computer simulation, coded in vb.net. If you could also suggest a numerical integragtion method I can possibly turn it into a vb.net program that can solve the system...Thanks!