vabsie
- 7
- 0
- TL;DR Summary
- I have 14 coupled ordinary differential equations. I am trying to increase the concentration of one of the ODEs by adding a sinusoidal equation that is at the same time "noisy". This noise I introduce follows a gaussian distribution with mean 0 and sigma that I vary in several experiments. I have not tried calculating for an analytical solution (not sure if even possible) but only used Python to solve the system. I don't know if I am doing it right. Any help is appreciated.
The Coupled ODE Model
Below are my coupled differential equations, where the only variable I try to meddle with is the ITMblood. The motivation here is if I try to increase ITMblood (in the next section I will show how I do it), at some concentration of ITMblood (most likely a very huge one) , the system below "collapses." That is, some of the concentrations just flattens out.
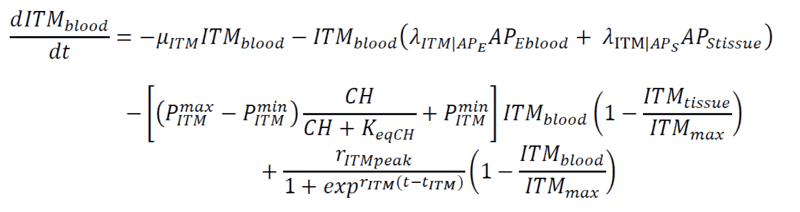
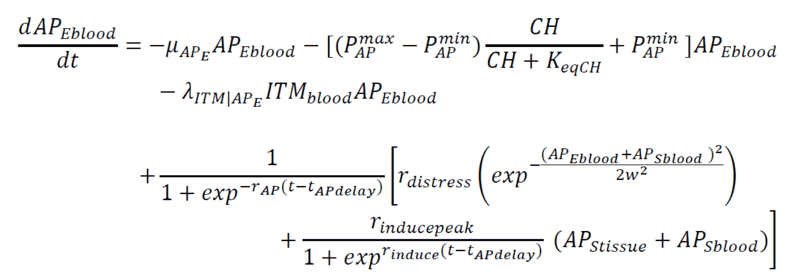
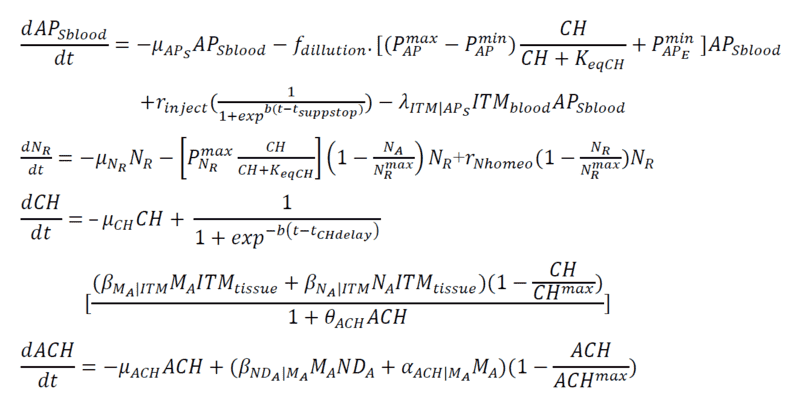
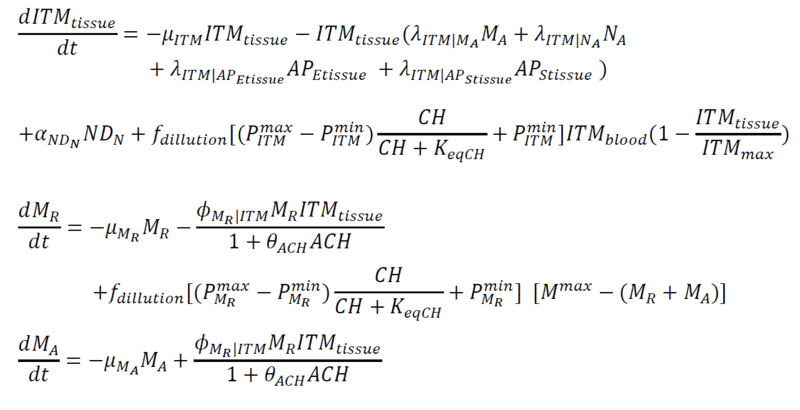
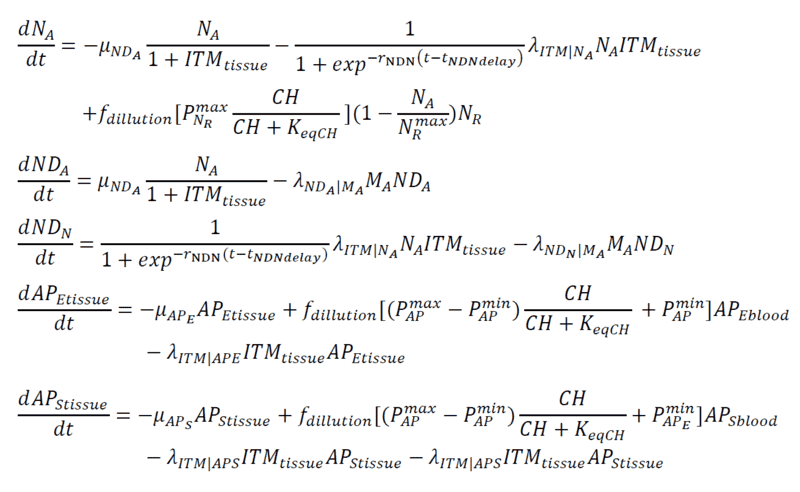
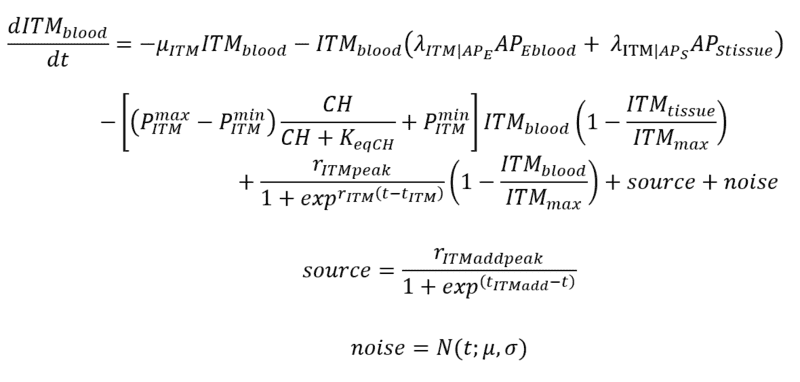
Introducing Noise to ITMblood
This is just a naive method that I am doing. So I just add a "source" of this ITMblood that follows a sinusoidal function which plateaus at rITMaddpeak and somehow starts "late" controlled by tITMadd. Then I add "noise" that simply follows a gaussian distribution with mu = 0 and sigma that I vary as part of my experiments.
Python Library Used to Solve the Equations
scipy.integrate.odeint
note: I have tried an existing library (sdeint) specifically designed for SDEs but for some reason, the solver just can't handle the system/blows up, even when I have not added noise yet (used this as sanity check).
Am I doing this right? Does this method of introducing noise make sense? Thank you very much! I can upload my notebooks if necessary.
Below are my coupled differential equations, where the only variable I try to meddle with is the ITMblood. The motivation here is if I try to increase ITMblood (in the next section I will show how I do it), at some concentration of ITMblood (most likely a very huge one) , the system below "collapses." That is, some of the concentrations just flattens out.
Introducing Noise to ITMblood
This is just a naive method that I am doing. So I just add a "source" of this ITMblood that follows a sinusoidal function which plateaus at rITMaddpeak and somehow starts "late" controlled by tITMadd. Then I add "noise" that simply follows a gaussian distribution with mu = 0 and sigma that I vary as part of my experiments.
Python Library Used to Solve the Equations
scipy.integrate.odeint
note: I have tried an existing library (sdeint) specifically designed for SDEs but for some reason, the solver just can't handle the system/blows up, even when I have not added noise yet (used this as sanity check).
Am I doing this right? Does this method of introducing noise make sense? Thank you very much! I can upload my notebooks if necessary.
Attachments
Last edited:




 .This is how I want to add the source term of ITMs so it won't increase linearly, and instead stop increasing at a maximum value. I hope that's clear?
.This is how I want to add the source term of ITMs so it won't increase linearly, and instead stop increasing at a maximum value. I hope that's clear? Sometimes, it gives out errors, sometimes it's ok.
Sometimes, it gives out errors, sometimes it's ok.