- 893
- 483
Trying this out for fun, and seeing if people find this stimulating or not. Feedback appreciated! There's only 3 problems, but I hope you'll get a kick out of them. Have fun!1. Springey Thingies:

Two damped, unforced springs are weakly coupled and obey the following equations of motion: $$\ddot{x}_a +\gamma \dot{x}_a + \omega_{0,a}^2 x_a + \beta^2 (x_b - x_a) = 0$$ $$\ddot{x}_b +\gamma \dot{x}_b + \omega_{0,b}^2 x_b - \beta^2 (x_b - x_a) = 0$$ You wish to measure the difference between the two springs' natural (undamped) resonant frequencies: ##\Delta = \omega_{0,b} - \omega_{0,a}##. Your measurement will be complicated by the coupling coefficient β and the damping coefficient γ. Design a simple procedure for measuring ##\Delta##.
Assume for simplicity that ##\omega_0 = \frac{1}{2}\left(\omega_{0,b} + \omega_{0,a}\right) = 1\mathrm{kHz}## and ##\gamma = 125\mathrm{s^{-1}}## are known exactly. You are also given that Δ is of order ##2\pi\times 1\mathrm{mHz}## and β is of order ##2\pi\times100\mathrm{mHz}##. The uncertainty in either spring's measured position is determined by the spring's initial conditions by $$\sigma_x = \left(2.6\times10^{-7}\right)\sqrt{x(0)^2 + \frac{\dot{x}(0)^2}{\omega_0^2 - \gamma^2 / 4}}$$
Your answer should include a set of times at which to measure the spring positions ##x_a## and ##x_b##, a formula for ##\Delta## in terms of these measurements, and a standard deviation on the value of ##\Delta##. Optimal answers should have uncertainty ##\sigma_\Delta \approx 2\pi \times 10\mathrm{\mu Hz}## with as little as two position measurements. Numerical and analytical methods are accepted so long as the results are valid!2. "Honey, I shrunk the error bars!"
You and your coworker Bob are studying a chemical reaction ##A + B \leftrightarrow C##. For this study, you vary the temperature of the mixture and record the concentration of species C: $$x = \frac{N_C}{N_A + N_B + N_C}$$ where ##N_A##, ##N_B##, and ##N_C## refer to the total number of each species A, B, and C respectively. For each temperature setting, you record a number (M) of measurements of ##N_A##, ##N_B##, ##N_C## (M measurements of each). Furthermore, you know that ##N_A##, ##N_B##, and ##N_C## are all Poisson distributed. You then calculate an average ##\mathrm{E}[x]## and standard error ##\sigma_{\mathrm{E}[x]}## for each set of measurements. Up to this point, everything makes sense.
Your coworker Bob comes up with a wacky idea. Bob re-defines the concentration (now called ##x'##) within a set of M measurements as follows: $$x'_i = \frac{N_{C,i}}{\mathrm{E}[N_A] + \mathrm{E}[N_B] + N_{C,i}} \; \; \mathrm{for} \; i=1,2,...,M$$ Bob argues that taking expectation values over ##N_A## and ##N_B## in the denominator eliminates extraneous noise. What's more, Bob has a mathematical proof that shows that ##\mathrm{Var}[x']\leq\mathrm{Var}[x]##. You make a bet with Bob: you collect 100 data sets, each consisting of M measurements, and compare the estimated standard error on the mean ##\sqrt{\frac{1}{M}\mathrm{Var}[x']}## (aka the "error bars" on the mean of each set of M measurements) with the observed standard deviation on the means of each of the 100 sets of measurements ##\sigma_{\mathrm{E}[x']}##. The data shows that ##\sigma_{\mathrm{E}[x']} > \sqrt{\frac{1}{M}\mathrm{Var}[x']}##, and more specifically that $${\sigma_{\mathrm{E}[x']}}=\sigma_{\mathrm{E}[x]}$$ This last result can be interpreted to mean there is no free lunch for Bob. Reproduce Bob's proof that ##\mathrm{Var}[x']\leq\mathrm{Var}[x]## and prove the "no free lunch" result ##\sigma_{\mathrm{E}[x']}=\sigma_{\mathrm{E}[x]}##.3. Pink, pink, you stink!
Consider the following bridge circuit, where the variable resistor sees "pink" noise (aka 1/f noise):
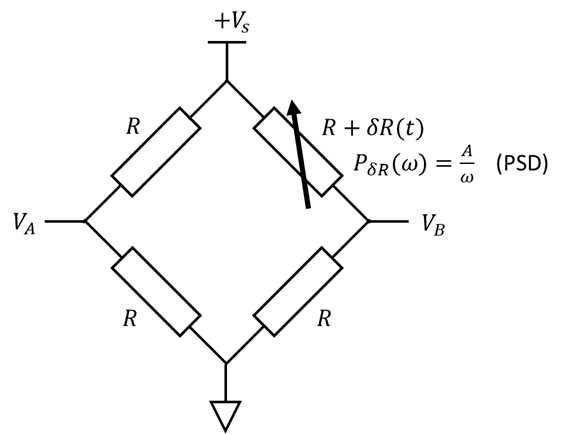
All 4 resistors have identical resistance on average, but the top right resistor fluctuates with a pink spectrum: $$P_{\delta R}(\omega) = \frac{A}{\omega}$$ where ##P_x(\omega)## is the power spectral density (PSD) of the function ##x(t)##. Each resistor also puts out thermal noise (Johnson-Nyquist noise). Find an expression in terms of the measured voltages ##V_A##, ##V_B##, and ##V_S## that is proportional to the fluctuating resistance ##\delta R## but is independent of thermal noise.
Some sample data is attached (filename is “pinkdatafinalfinal.csv”), where each voltage (##V_A##,##V_B##, and ##V_S##) is reported versus time in a CSV format. Extract the constant A as defined above and state your uncertainty on A, given ##R = 1\mathrm{\Omega}##. My solution has uncertainty on the order of ##1 \times 10^{-8} \mathrm{\mathrm{\Omega^2}}##. There are many methods for tackling this problem and some give higher precision than others.
Two damped, unforced springs are weakly coupled and obey the following equations of motion: $$\ddot{x}_a +\gamma \dot{x}_a + \omega_{0,a}^2 x_a + \beta^2 (x_b - x_a) = 0$$ $$\ddot{x}_b +\gamma \dot{x}_b + \omega_{0,b}^2 x_b - \beta^2 (x_b - x_a) = 0$$ You wish to measure the difference between the two springs' natural (undamped) resonant frequencies: ##\Delta = \omega_{0,b} - \omega_{0,a}##. Your measurement will be complicated by the coupling coefficient β and the damping coefficient γ. Design a simple procedure for measuring ##\Delta##.
Assume for simplicity that ##\omega_0 = \frac{1}{2}\left(\omega_{0,b} + \omega_{0,a}\right) = 1\mathrm{kHz}## and ##\gamma = 125\mathrm{s^{-1}}## are known exactly. You are also given that Δ is of order ##2\pi\times 1\mathrm{mHz}## and β is of order ##2\pi\times100\mathrm{mHz}##. The uncertainty in either spring's measured position is determined by the spring's initial conditions by $$\sigma_x = \left(2.6\times10^{-7}\right)\sqrt{x(0)^2 + \frac{\dot{x}(0)^2}{\omega_0^2 - \gamma^2 / 4}}$$
Your answer should include a set of times at which to measure the spring positions ##x_a## and ##x_b##, a formula for ##\Delta## in terms of these measurements, and a standard deviation on the value of ##\Delta##. Optimal answers should have uncertainty ##\sigma_\Delta \approx 2\pi \times 10\mathrm{\mu Hz}## with as little as two position measurements. Numerical and analytical methods are accepted so long as the results are valid!2. "Honey, I shrunk the error bars!"
You and your coworker Bob are studying a chemical reaction ##A + B \leftrightarrow C##. For this study, you vary the temperature of the mixture and record the concentration of species C: $$x = \frac{N_C}{N_A + N_B + N_C}$$ where ##N_A##, ##N_B##, and ##N_C## refer to the total number of each species A, B, and C respectively. For each temperature setting, you record a number (M) of measurements of ##N_A##, ##N_B##, ##N_C## (M measurements of each). Furthermore, you know that ##N_A##, ##N_B##, and ##N_C## are all Poisson distributed. You then calculate an average ##\mathrm{E}[x]## and standard error ##\sigma_{\mathrm{E}[x]}## for each set of measurements. Up to this point, everything makes sense.
Your coworker Bob comes up with a wacky idea. Bob re-defines the concentration (now called ##x'##) within a set of M measurements as follows: $$x'_i = \frac{N_{C,i}}{\mathrm{E}[N_A] + \mathrm{E}[N_B] + N_{C,i}} \; \; \mathrm{for} \; i=1,2,...,M$$ Bob argues that taking expectation values over ##N_A## and ##N_B## in the denominator eliminates extraneous noise. What's more, Bob has a mathematical proof that shows that ##\mathrm{Var}[x']\leq\mathrm{Var}[x]##. You make a bet with Bob: you collect 100 data sets, each consisting of M measurements, and compare the estimated standard error on the mean ##\sqrt{\frac{1}{M}\mathrm{Var}[x']}## (aka the "error bars" on the mean of each set of M measurements) with the observed standard deviation on the means of each of the 100 sets of measurements ##\sigma_{\mathrm{E}[x']}##. The data shows that ##\sigma_{\mathrm{E}[x']} > \sqrt{\frac{1}{M}\mathrm{Var}[x']}##, and more specifically that $${\sigma_{\mathrm{E}[x']}}=\sigma_{\mathrm{E}[x]}$$ This last result can be interpreted to mean there is no free lunch for Bob. Reproduce Bob's proof that ##\mathrm{Var}[x']\leq\mathrm{Var}[x]## and prove the "no free lunch" result ##\sigma_{\mathrm{E}[x']}=\sigma_{\mathrm{E}[x]}##.3. Pink, pink, you stink!
Consider the following bridge circuit, where the variable resistor sees "pink" noise (aka 1/f noise):
All 4 resistors have identical resistance on average, but the top right resistor fluctuates with a pink spectrum: $$P_{\delta R}(\omega) = \frac{A}{\omega}$$ where ##P_x(\omega)## is the power spectral density (PSD) of the function ##x(t)##. Each resistor also puts out thermal noise (Johnson-Nyquist noise). Find an expression in terms of the measured voltages ##V_A##, ##V_B##, and ##V_S## that is proportional to the fluctuating resistance ##\delta R## but is independent of thermal noise.
Some sample data is attached (filename is “pinkdatafinalfinal.csv”), where each voltage (##V_A##,##V_B##, and ##V_S##) is reported versus time in a CSV format. Extract the constant A as defined above and state your uncertainty on A, given ##R = 1\mathrm{\Omega}##. My solution has uncertainty on the order of ##1 \times 10^{-8} \mathrm{\mathrm{\Omega^2}}##. There are many methods for tackling this problem and some give higher precision than others.
 Here are some hints if y'all are still interested!
Here are some hints if y'all are still interested!